Again, we will start with preparing the data in native space. Do the DTI preprocessing up to the tensor estimation, so you can start tracking fibers. The next step is to select the tract you are interested in, in the DTI > Fibers Table menu. Then, transform the fibers to ACPC or TAL space, as explained in 1.8.3.
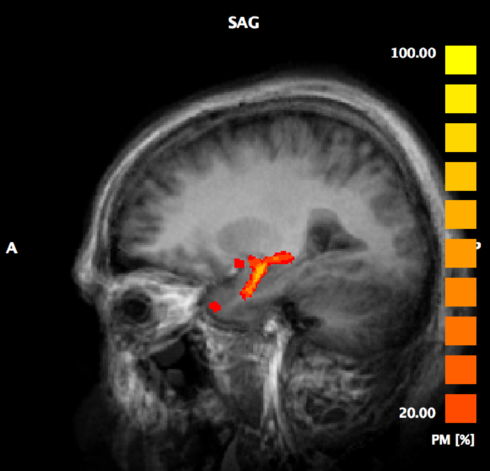
Next, go to DTI > Spatial transformations and click the button Fibers -> VMR. This will project the 3-D fiber reconstruction onto the 2-D VMR slices. The result is a volume of interest (VOI) on the VMR. As explained earlier, if you add a VMP in Talairach space, you can extract the MD/ADC values from that VOI.

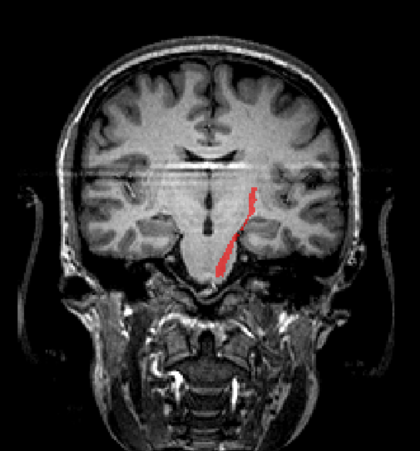
Ofcourse we could overlay the tracts from all subjects on a TAL vmr, but a nicer option is to create probability maps of the tracts across subjects. To do this, convert the tracts from all subjects into TAL space, and use the naming convention <subject>_tractname for each voi, eg CG_CST. Add the vois of all subjects into one voi file, by using the Add function in the VOI analysis dialog.
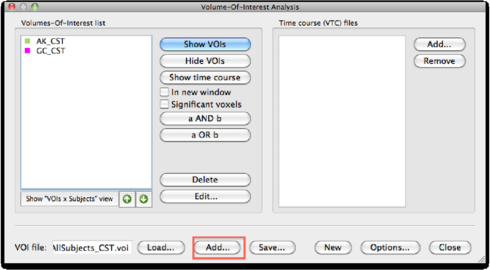
Then, click Options > VOI functions > Probability Maps, Create. Make sure you see only the tract name and not all subject names. You can use the Naming Convention radio buttons to change. Set the map resolution to 1x1x1 and click GO. The result is a VMP showing overlapping tracts. The colors indicate in what degree the tracts are overlapping. The percentage value is calculated according to 100 * (number of times a tract overlaps)/(number of subjects).