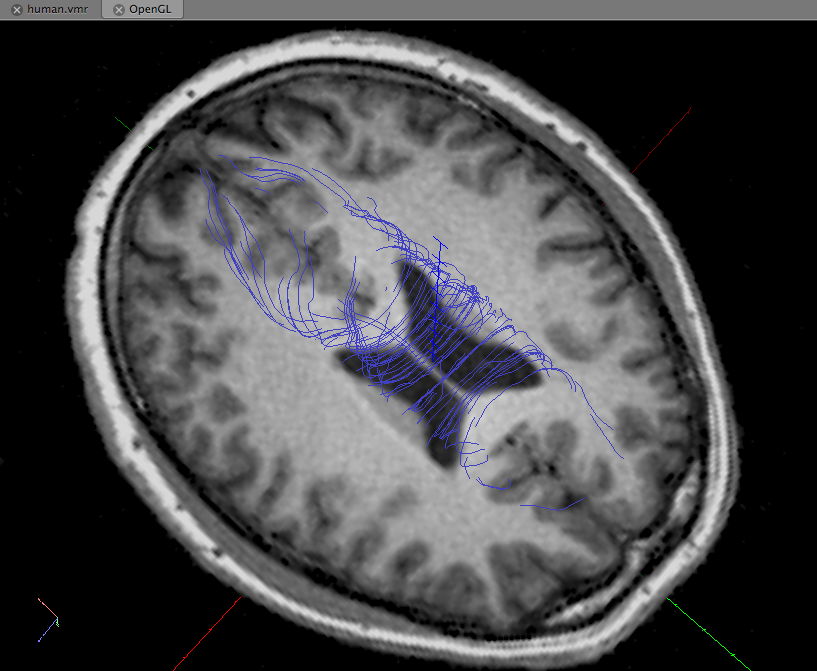
Convert MRtrix fiber files to BrainVoyager fiber files
- Details
- Category: Interaction With Other Software
- Last Updated: 16 April 2018
- Published: 16 April 2018
- Hits: 3333
Data preprocessing
This document was created with
• BrainVoyagerQX 2.2 build 1635 64-bit on Mac OSX 10.6
• BrainVoyagerNiftipluginv1.08.180610onMacOSX10.6(includedinthe default installation of BVQX 2.2)
• MRTrix 0.2.8 on Debian Lenny 64-bit
1. Create a DMR project in BVQX. Use the Nifti converter to export it to Nifti. Do not transform the data.
2. Create a VMR project in BVQX. Use the Nifti converter to export it to Nifti. Do not transform the data.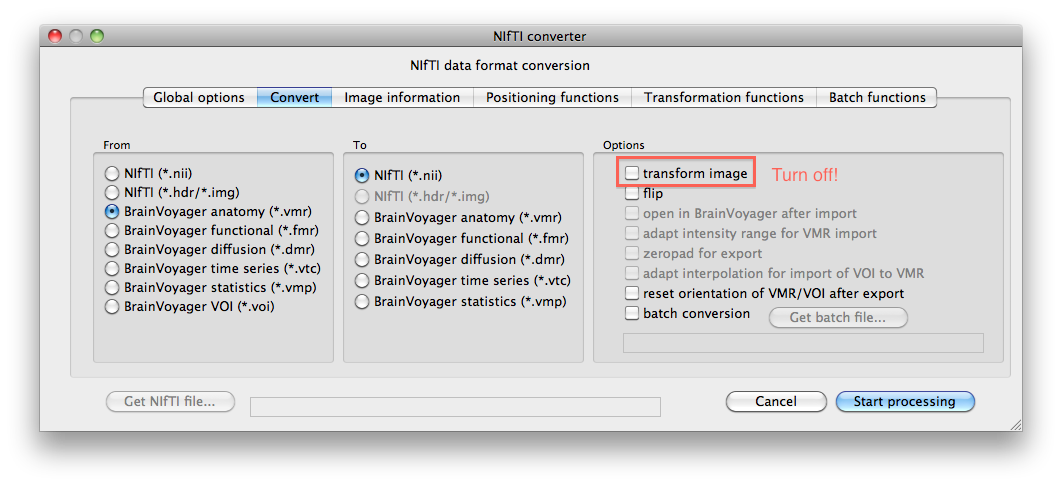
3. Create a region of interest in BVQX. Save it as a *.voi file. Use the Nifti converter to export it to Nifti. Do not transform the data.
4. Create the gradient file in BVQX, including the b-values. Save it as a grb file. In this example, I needed to flip the y and z directions. Open the *.grb file with a text editor. Remove all the header information but be sure to keep the gradient directions (the 4 columns with gradient direc- tions and b-values). I used matlab to invert the y and z directions, that is:
>> importdata(’grad.txt’);
>> grad(:,2) = -grad(:,2);
>> grad(:,3) = -grad(:,3);
>> dlmwrite(’grad_yzflip.b’,grad,’delimiter’,’ ’);
5. Now we‘re ready for MRTrix. Use the standard pipeline. Go to the direc- tory where you have saved the Nifti files of the anatomy, DMR data and VOI(s).
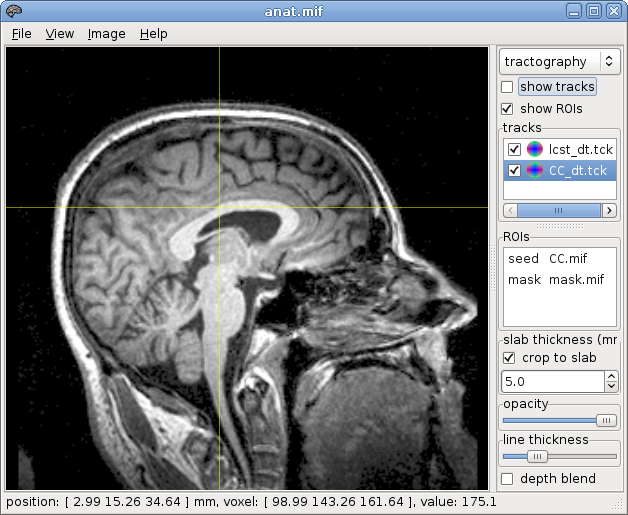
~$ mrconvert anat.nii anat.mif
̃
~$ mrconvert dwi.nii dwi.mif
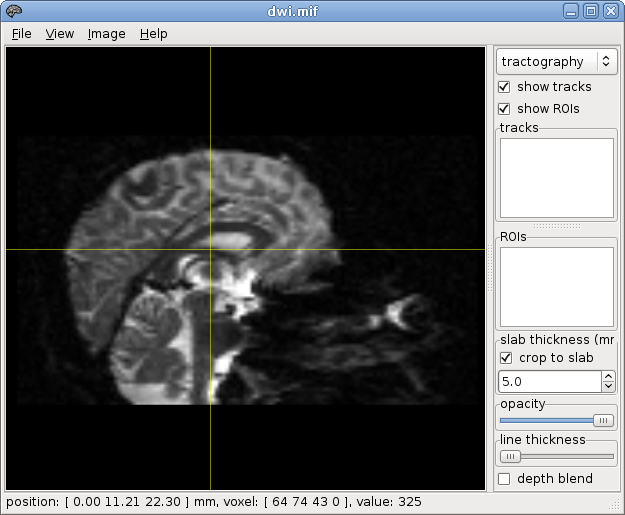
~$ mrconvert voi.nii voi.mif
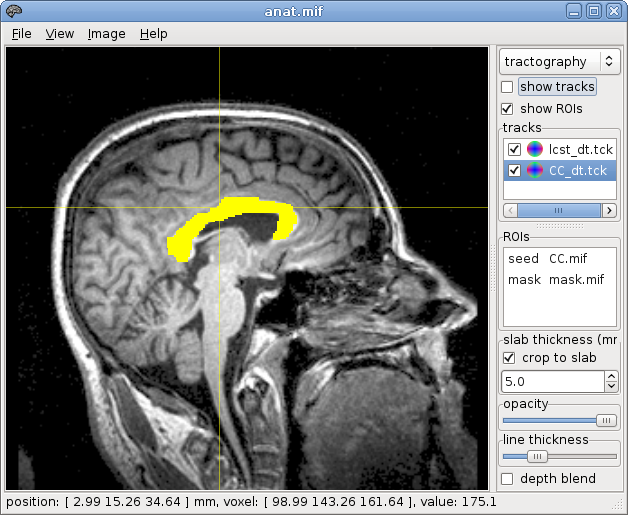
create a mask
~$ mrconvert dwi.mif -coord 3 0 - | threshold - - | median3D - - | median3D - mask.mif
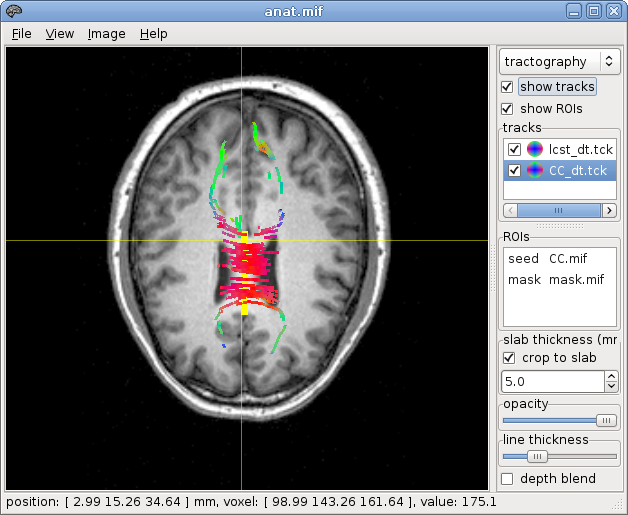
fiber tracking from the VOI:
~$ streamtrack DT_STREAM dwi.mif -seed voi.mif -mask mask.mif -grad grad_yzflip.b cst_dt.tck
et voila !
Converting the MRtrix fiber file to BrainVoyager fiber file
mrtrix2bvfiber.m
MRtrix software and read_mrtrix_tracks.m by J-D Tournier, Brain Research Institute, Melbourne,
Australia, http://www.brain.org.au/software/
usage:
>> fbr = mrtrix2bvfiber(xdim,ydim,zdim);
where xdim: anterior-posterior dimension of VMR(!)
ydim: inferior-superior dimension
zdim: left-right dimension
or alternatively
>> fbr = mrtrix2fiber;
requires BVQXtools package from
http://support.brainvoyager.com/available-tools/52-matlab-tools-bvxqtools.html