Masking the VTC
- Details
- Category: Creating Masks
- Last Updated: 11 April 2018
- Published: 11 April 2018
- Hits: 9119
In some cases, one wants to exclude specific areas from the functional data itself. There is no option to exclude something from the VTC itself within BrainVoyager, but there is an easy workaround to create a mask based on the properties of VTC.
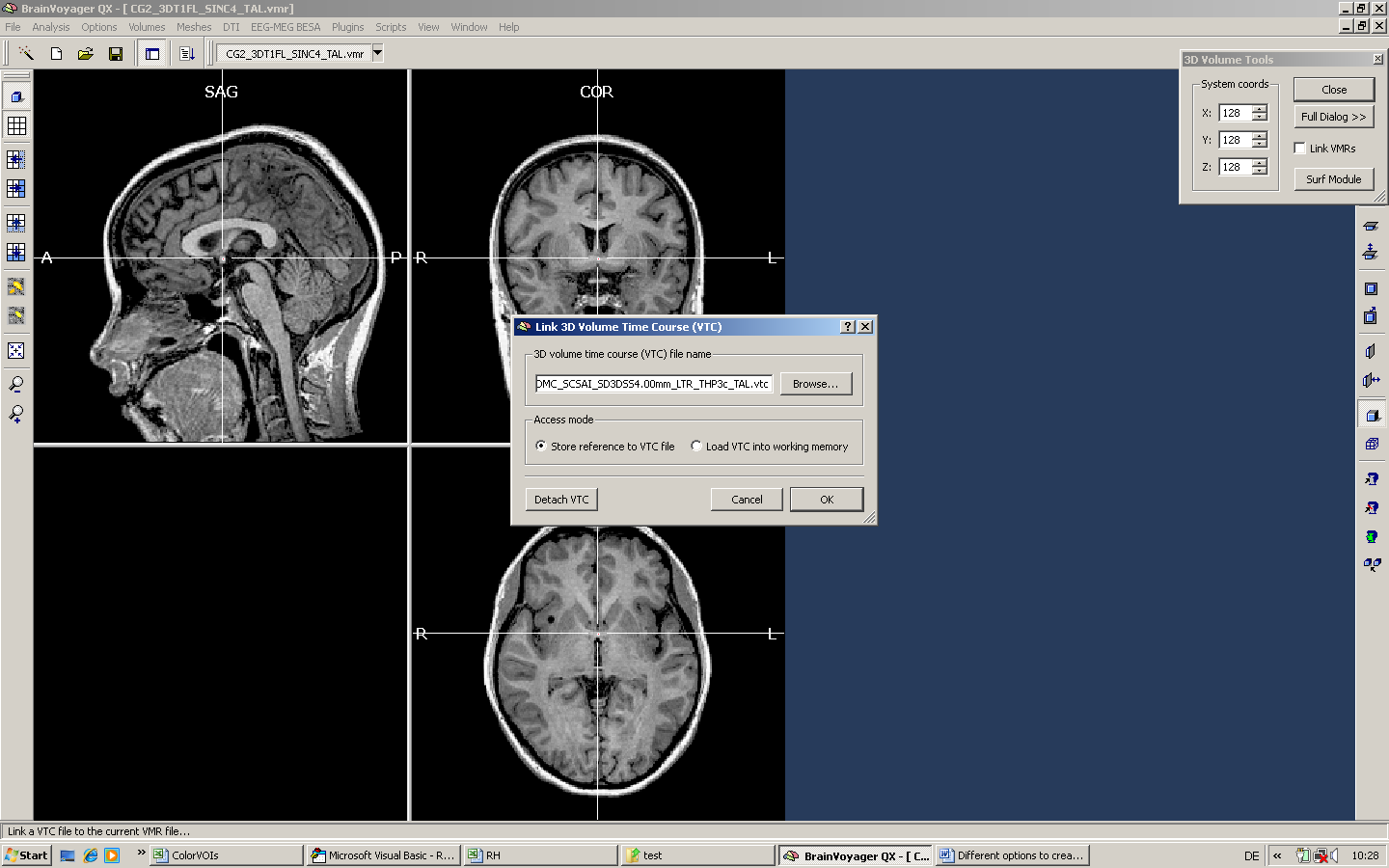
First, we have to link a VTC file to the VMR.
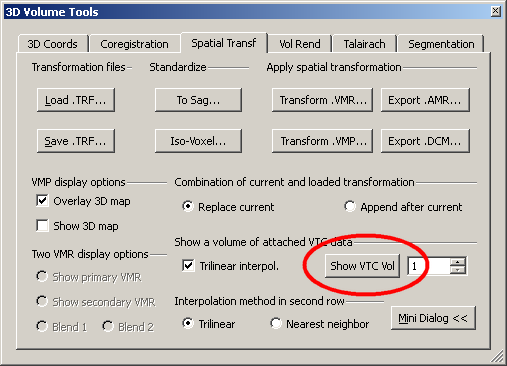
We visualize the VTC by using the “Show VTC vol” button on the Spatial Transf tab of the 3D volume tool. Here, the first VTC volume is displayed.
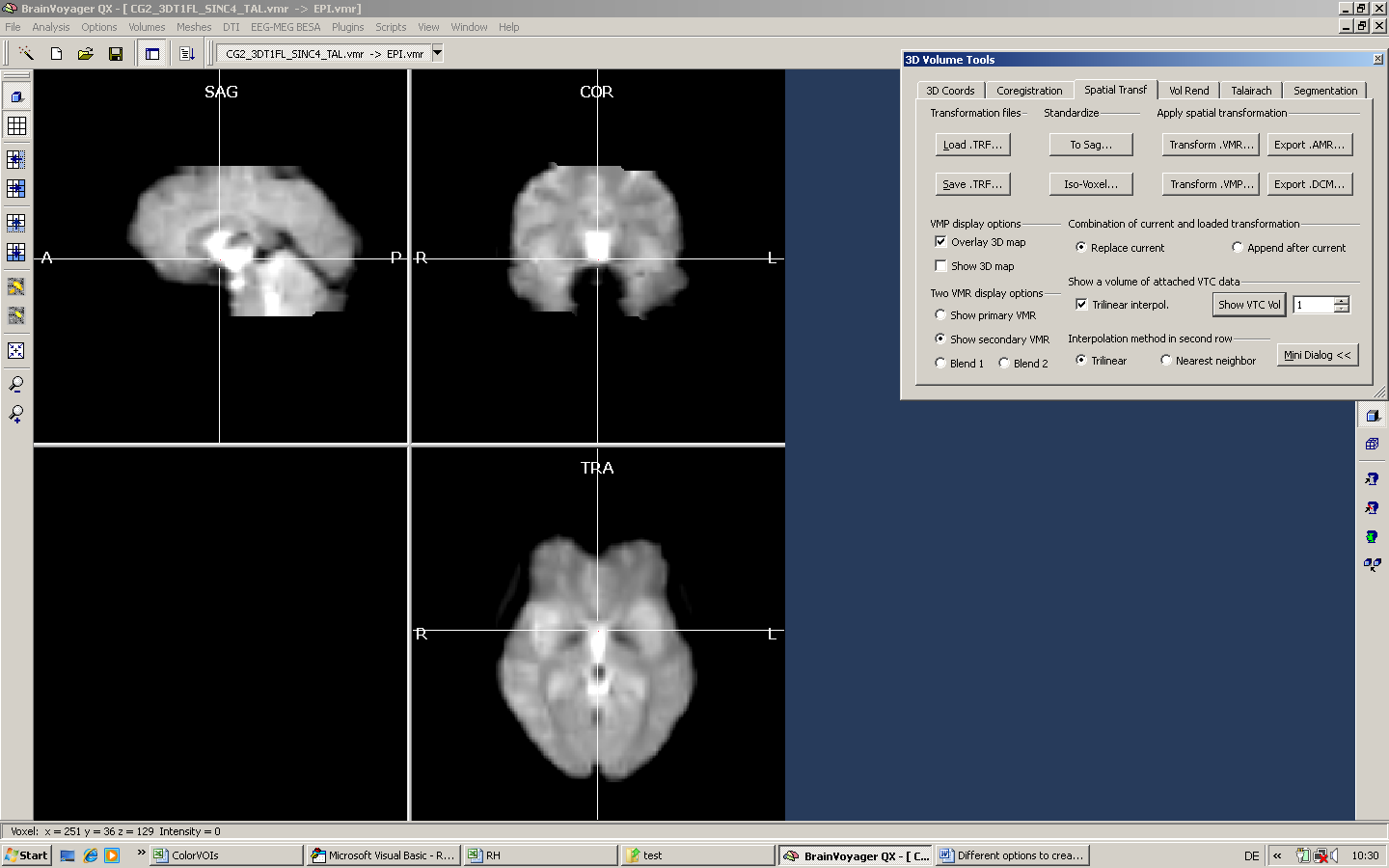
Now, we first have to save the VTC – which is temporarily displayed as “EPI.vmr” in the VMR format. To do so, we use the option “save seconday VMR” in the File menu.
We save the first VTC volume as VTC1.vmr and then load this new VTC1.vmr.
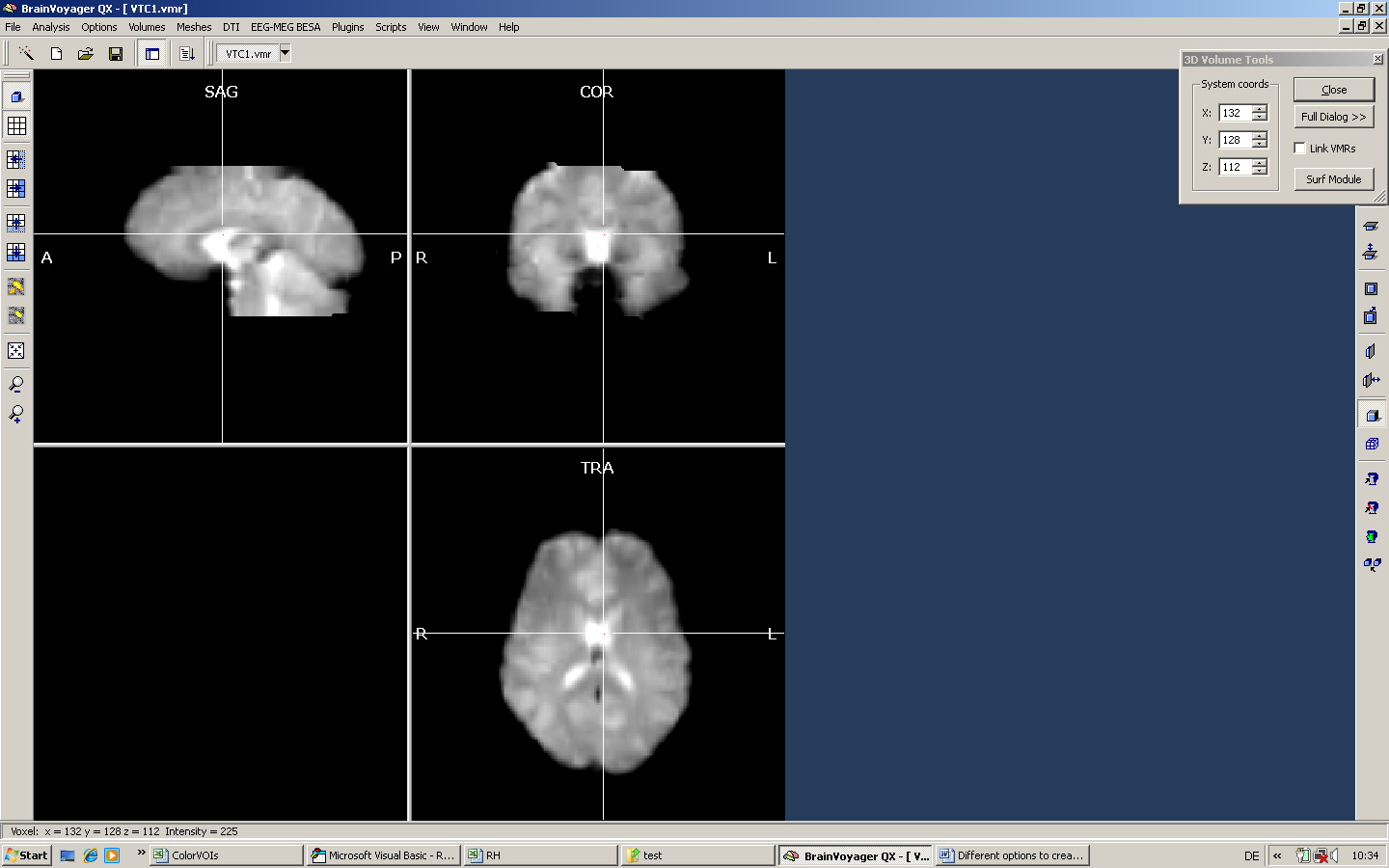
One idea may be to get rid of the ventricles in our mask. Moving the mouse over the ventricles shows that the ventricles are very bright (intensity value of 225). Now we could follow two different strategies. We could a) either mark the ventricles first and reload the non-marked area afterwards or b) mark everything except the ventricles. Here, we show the first approach.
a) We position the mouse inside the ventricles and set a very high intensity for the Min and Max values (here, we use just a single value - 225).
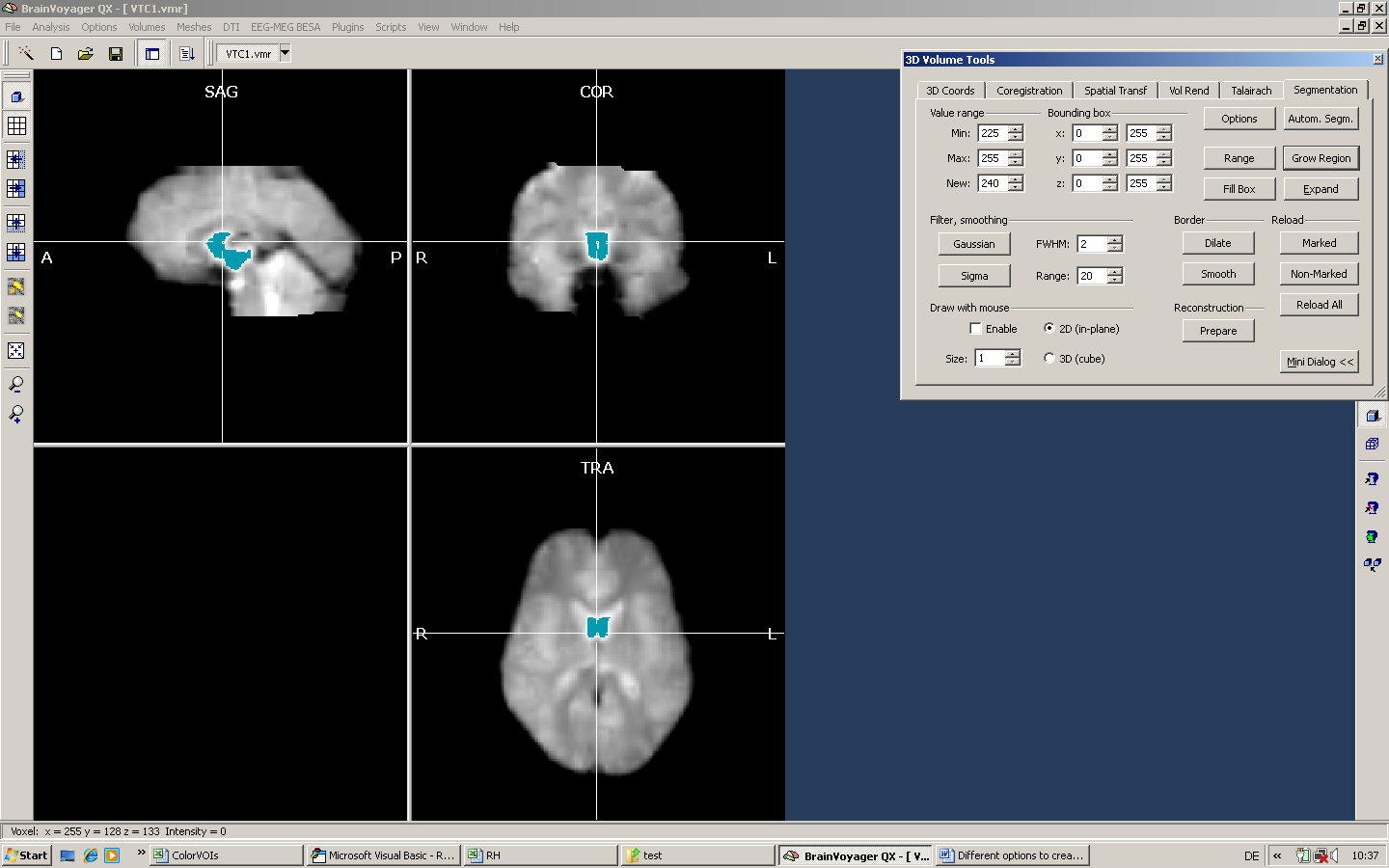
Only a part of the ventricles has been marked. By using the expansion tool, we can increase the size of the marked area.
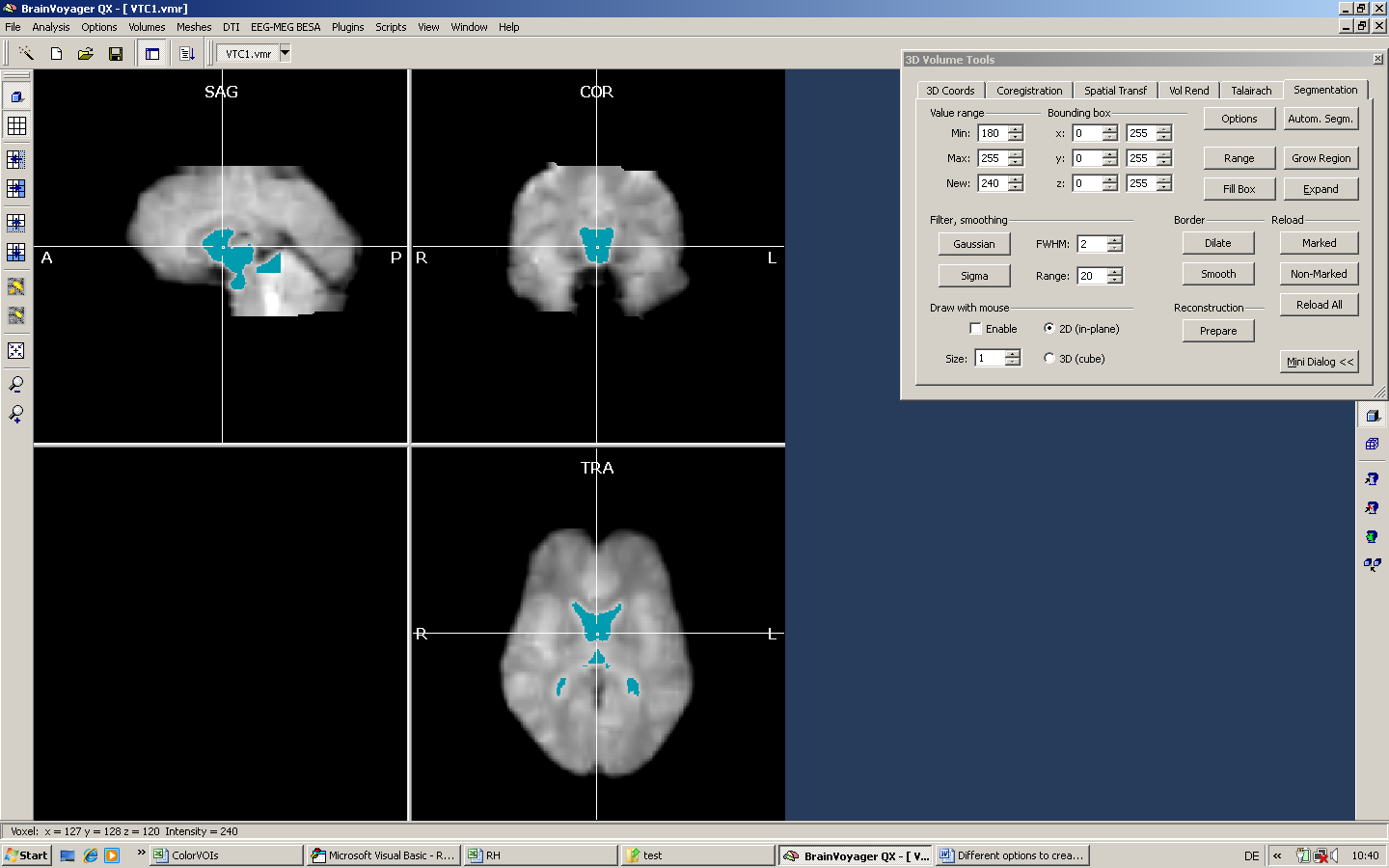
Depending on the specific distribution of intensities in the current dataset, we have to adapt the minimum value more or less. As an additional approach, we may also use manual drawing to mark fine structures.
The drawing tool can be enabled on the Segmentation tab.
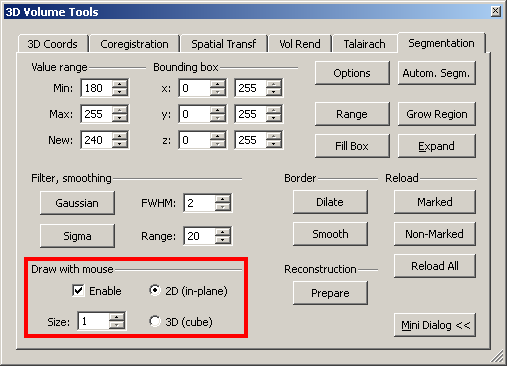
Depending on the size of the region to be manually marked, one may adjust the mouse size.
We use the expand button to increase the marked area and use to “Reload Non-Marked” button to get rid of the marked area.
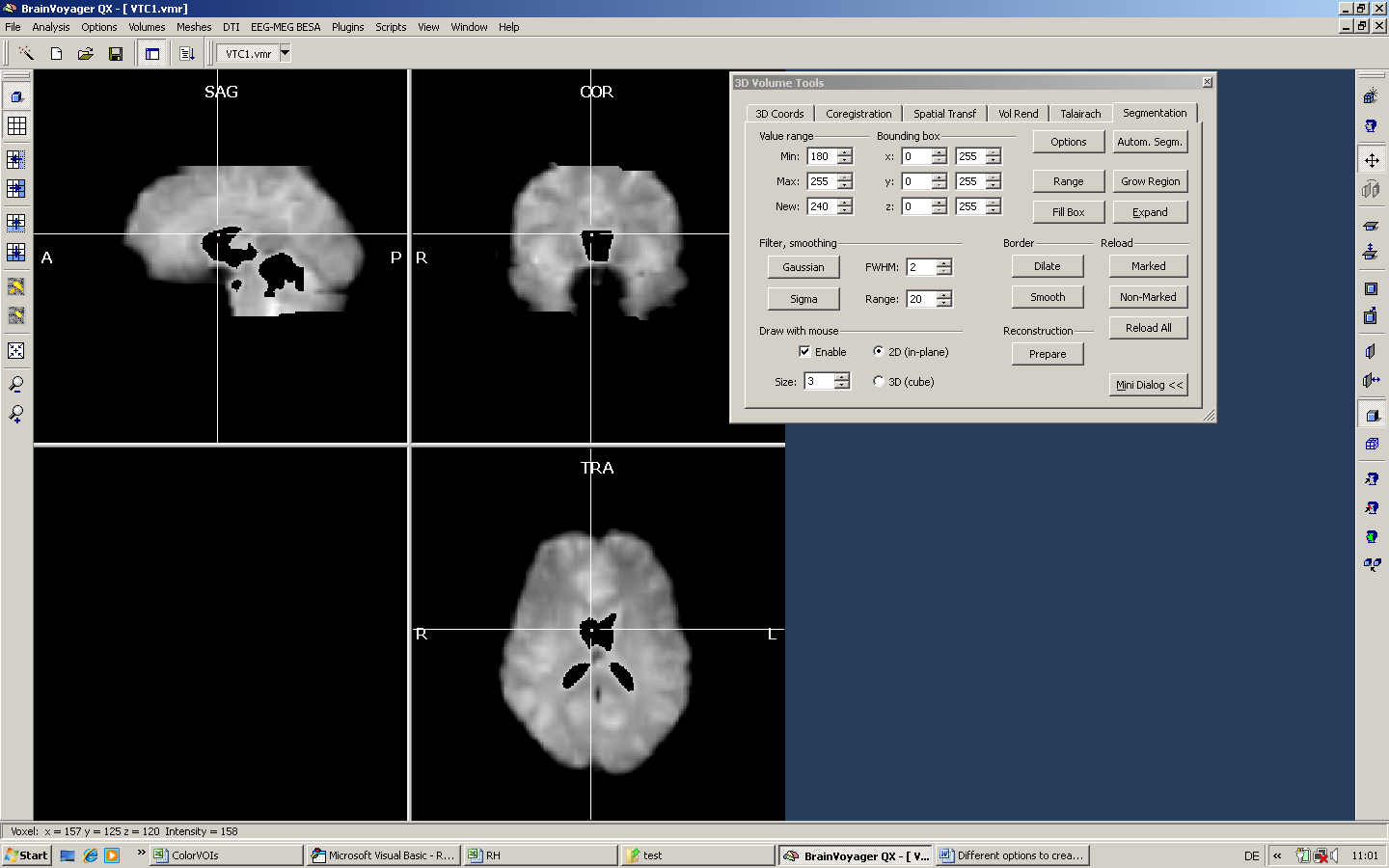
The new VMR should be saved before applying the next step.
To finally create the mask (after some manual corrections within the VTC1.vmr), we use exactly the same technique as performed in the first step. First, we mark all the voxels in the VMR using region growing / expansion. Due to the fact that we basically want to include everything larger than zero in the mask, we can use a quite large intensity range.
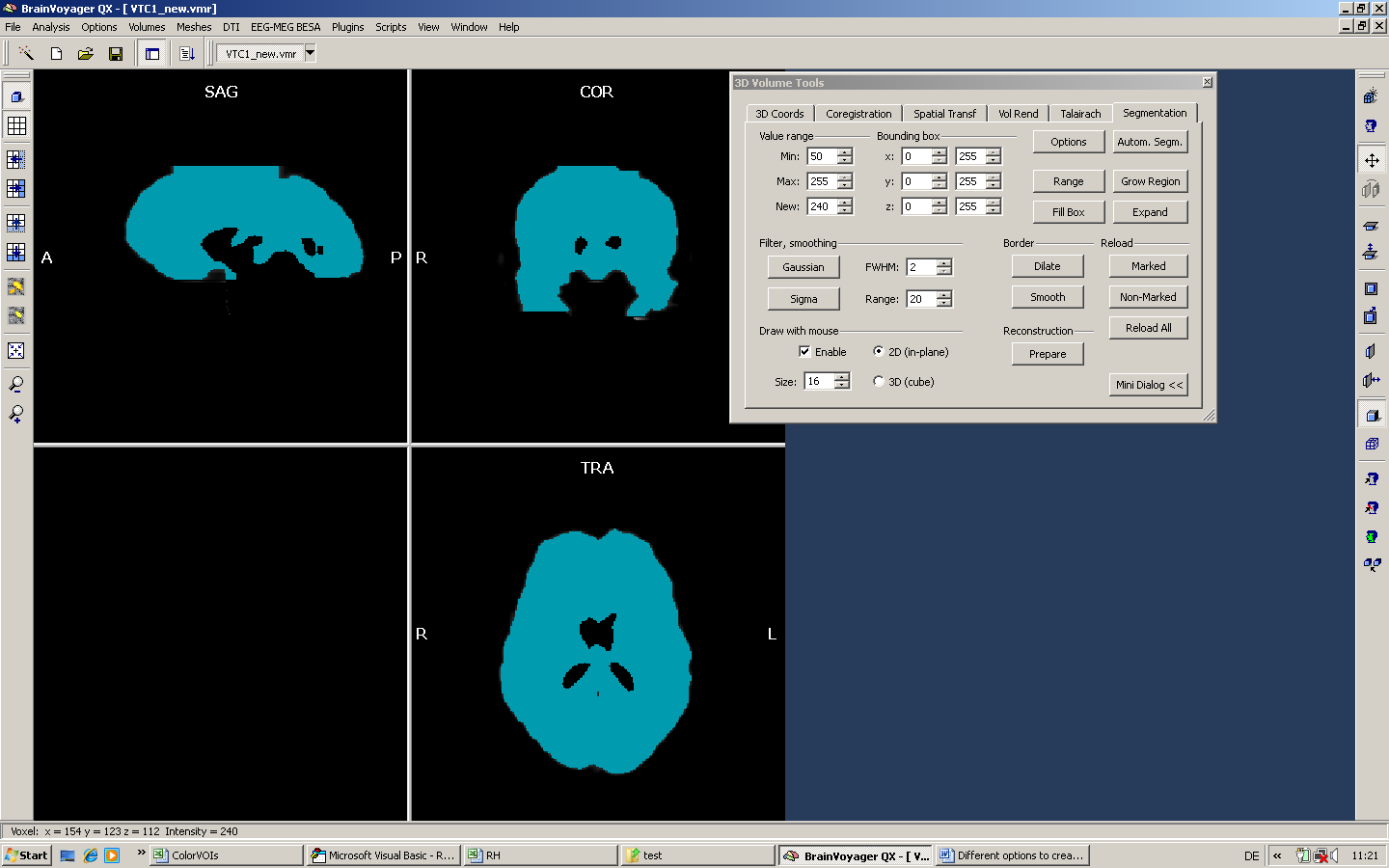
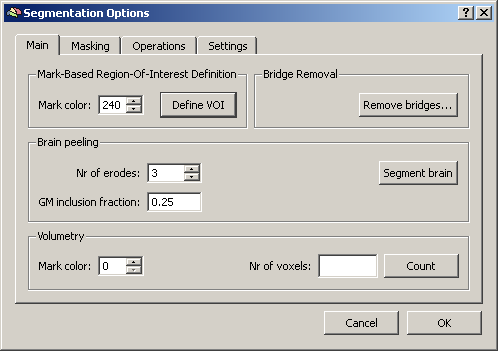
Second, we save the marked region as VOI file in the Options of the Segmentation tab.
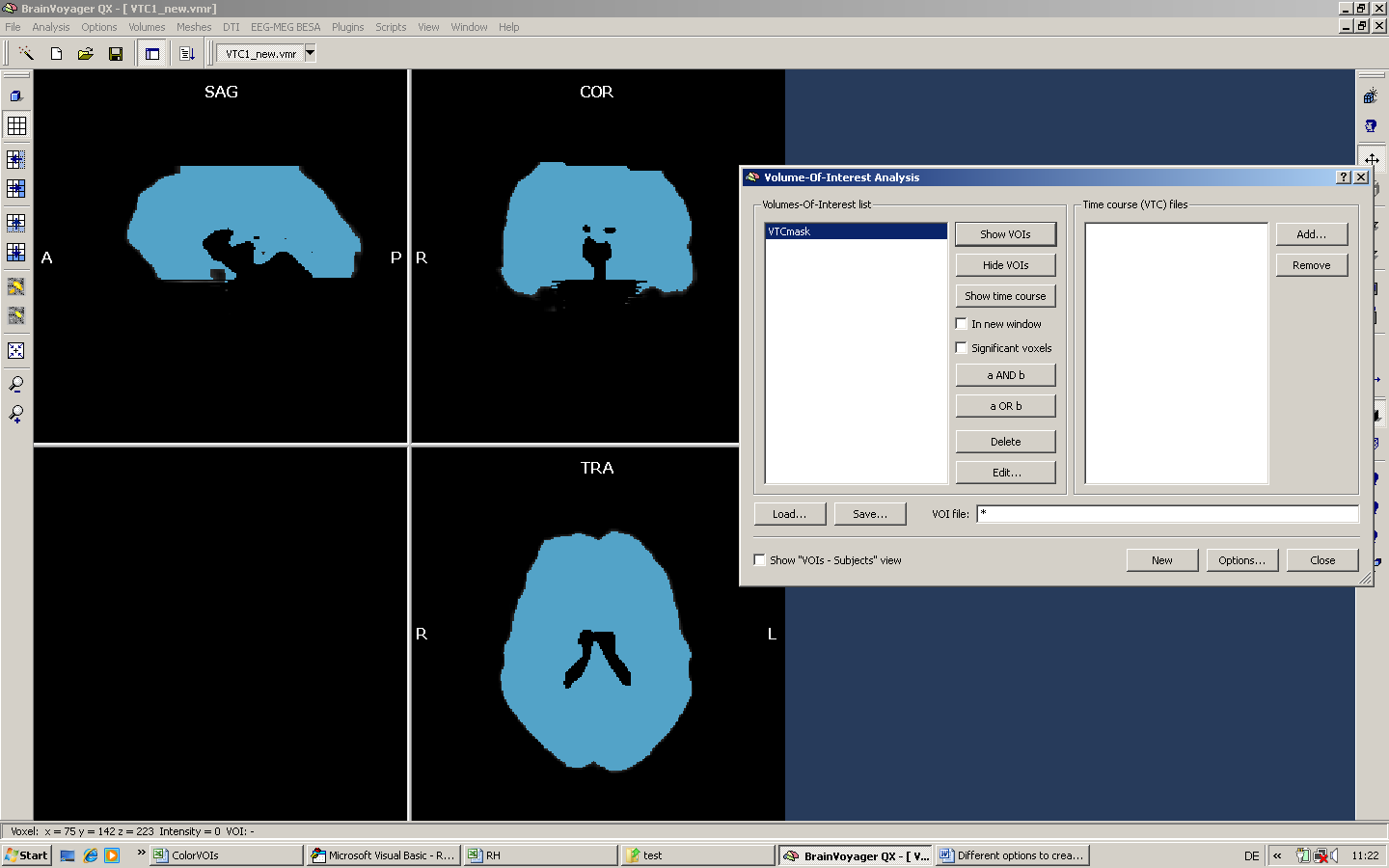
Third, we create a mask on the basis of the VOI.
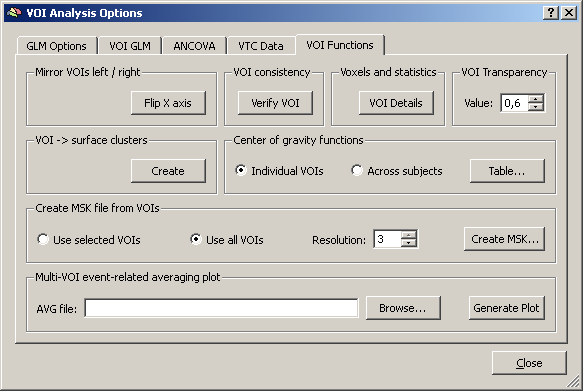
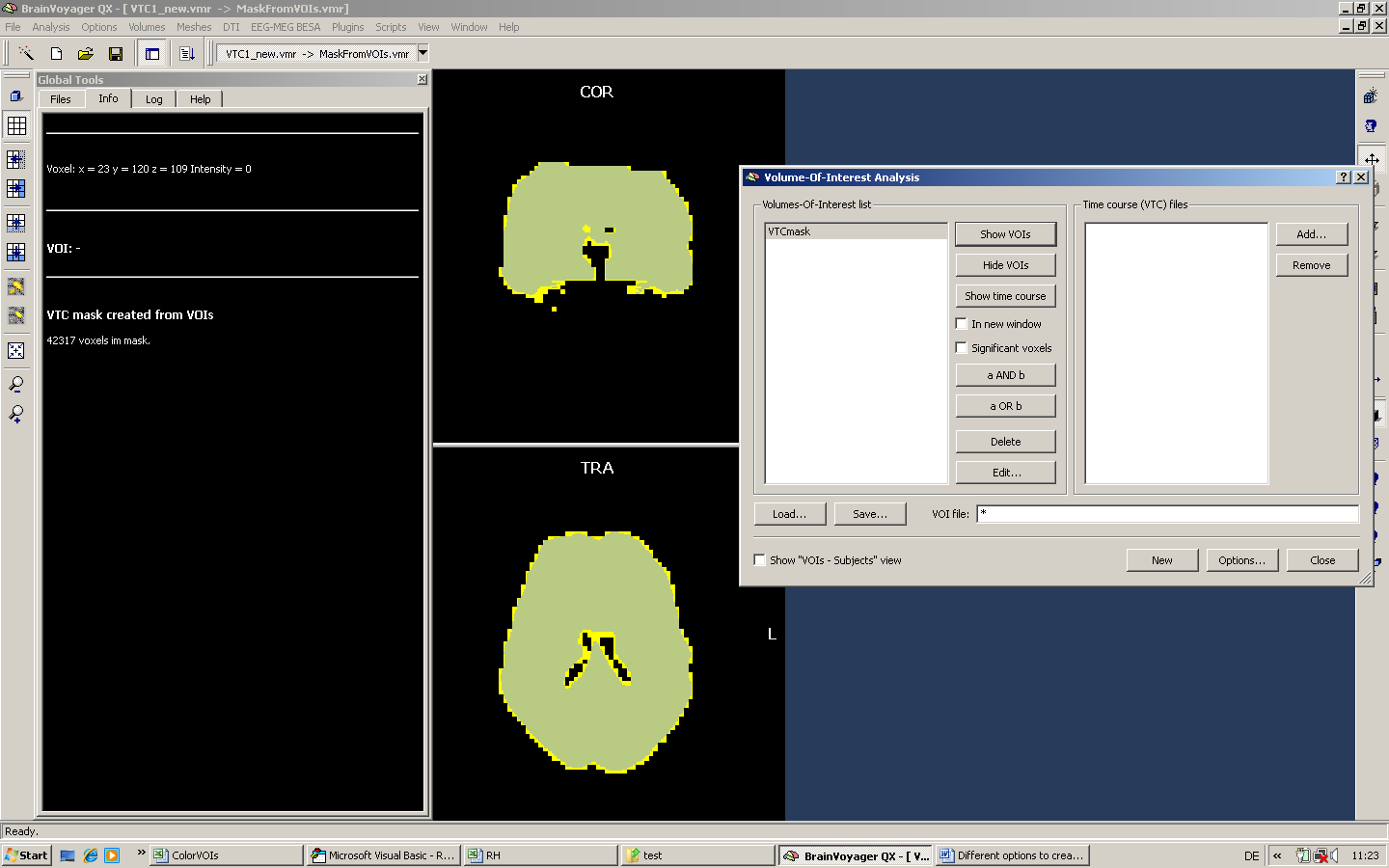
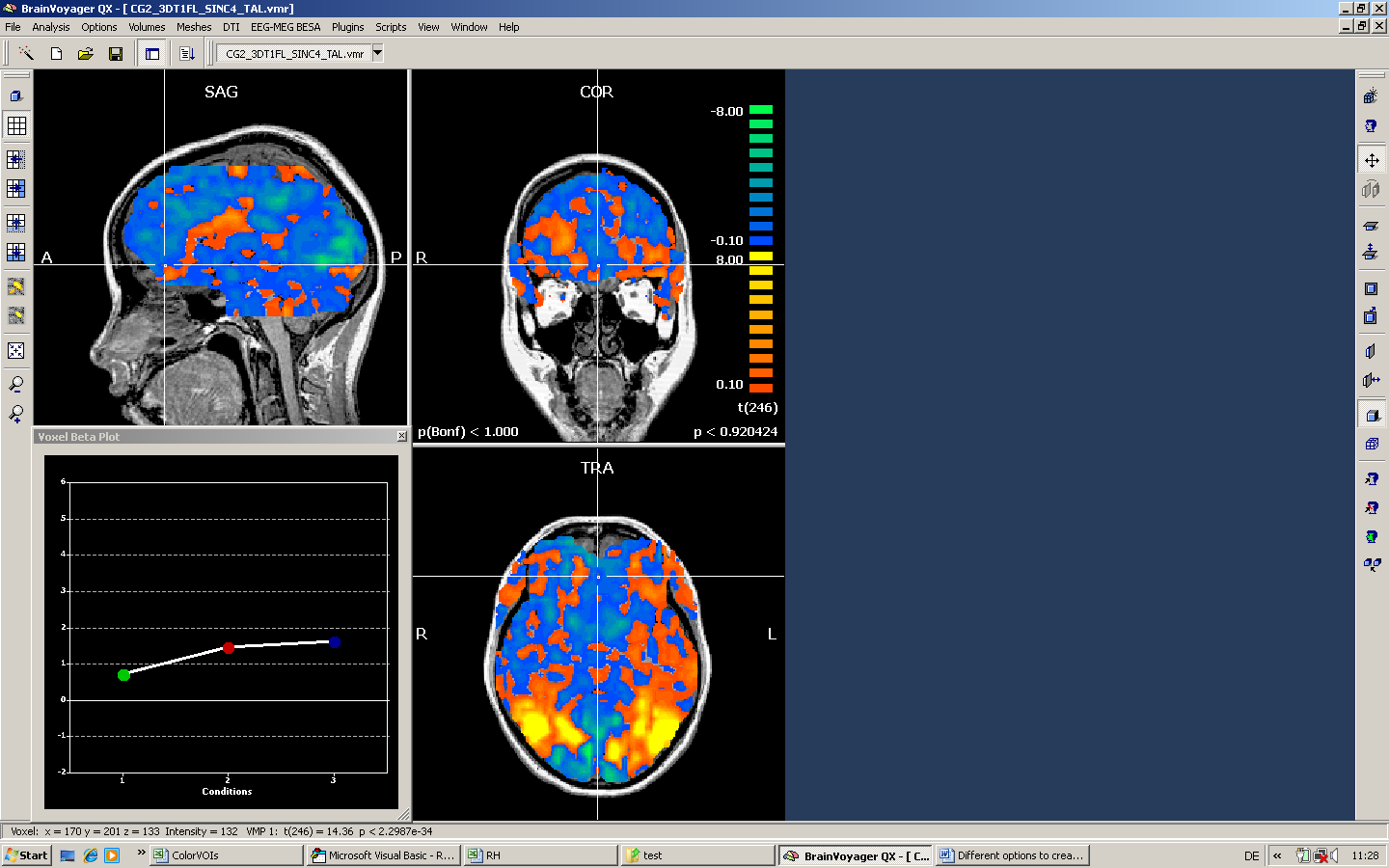
We apply the mask for a GLM analysis and can nicely test that the mask has been properly used by setting the minimal threshold to a very low value (but slightly above zero).
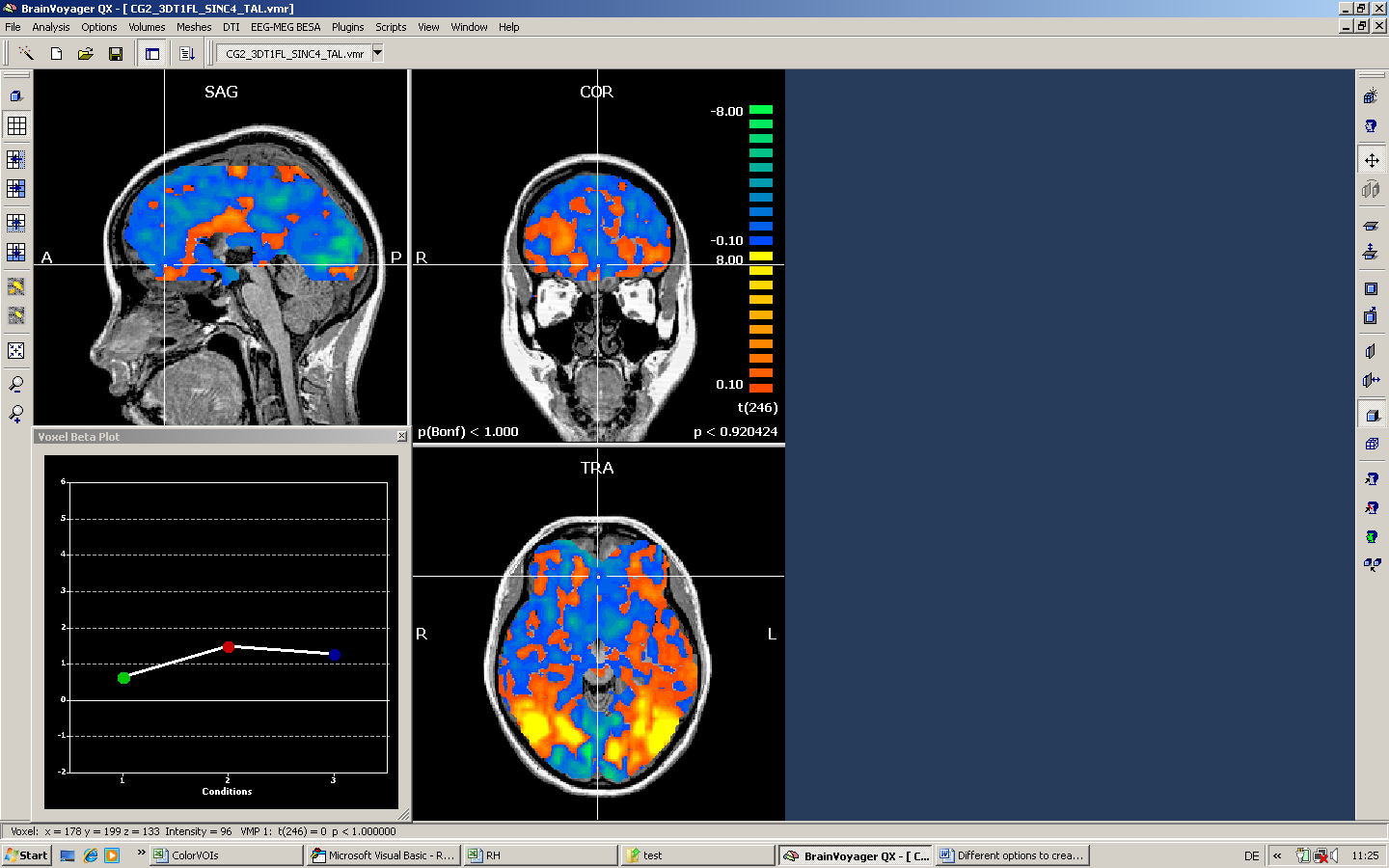
The following screenshot shows the corresponding result when no mask is used.