Use of probabilistic fiber atlases
- Details
- Category: Fiber Tracking
- Last Updated: 16 April 2018
- Published: 16 April 2018
- Hits: 4673
This page describes how to import the JHU white matter labels and probabilistic fiber atlas, available with FSL, into BrainVoyager. For information see http://www.fmrib.ox.ac.uk/fsl/data/atlas-descriptions.html#wm
You need a working FSL installation to do this.
In the ICBM-DTI-81 white-matter labels atlas, 50 white matter tract labels were created by hand segmentation of a standard-space average of diffusion MRI tensor maps from 81 subjects; mean age 39 (18:59), M:42, F: 39. The diffusion data was kindly provided by the ICBM DTI workgroup.
In the JHU white-matter tractography atlas, 20 structures were identified probabilistically by averaging the results of running deterministic tractography on 28 normal subjects (mean age 29, M:17, F:11).
You can find the atlases in $FSLDIR/data/atlases/JHU. In this directory you will also find xml files describing the content of the files.
To convert these into BrainVoyager files, do the following; First, copy the files you're interested in to a different location on your harddisk. Then, do at the terminal, for example
~$ fslswapdim JHU-ICBM-tracts-maxprob-thr0-1mm -x -y z JHU-ICBM-tracts-maxprob-thr0-1mm_swapxy
~$ gunzip *swapxy.nii.gz
now, you have the file JHU-ICBM-tracts-maxprob-thr0-1mm_swapxy.nii in the correct orientation to import into BrainVoyager. Import it with the Nifti converter as a VMR, using the default options. Once you have the file as a VMR, you will not see the data in the file, because intensities are in the range 1-20.
![]()
Repeat the import procedure, including the x-y swap for the file MNI152_T1_1mm_brain.nii.gz. This file, like the tract file is in MNI space, but we want to go to TAL space. We can use the T1 brain for the TAL transformation and apply the transformations to the fiber tract files.
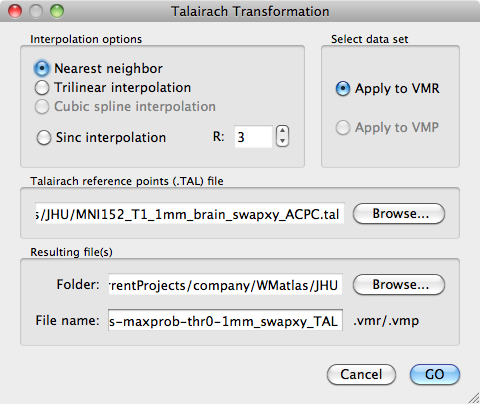
Open the MNI152_T1_1mm_brain_swapxy.vmr file and convert it to TAL. Apply the TAL transformations to JHU-ICBM-tracts-maxprob-thr0-1mm_swapxy.vmr. Be sure to use Nearest Neighbour interpolation to not change the voxel intensities in the file!
We will now continue to convert the VMR into VOI files. Open JHU-ICBM-tracts-maxprob-thr0-1mm_swapxy_TAL.vmr
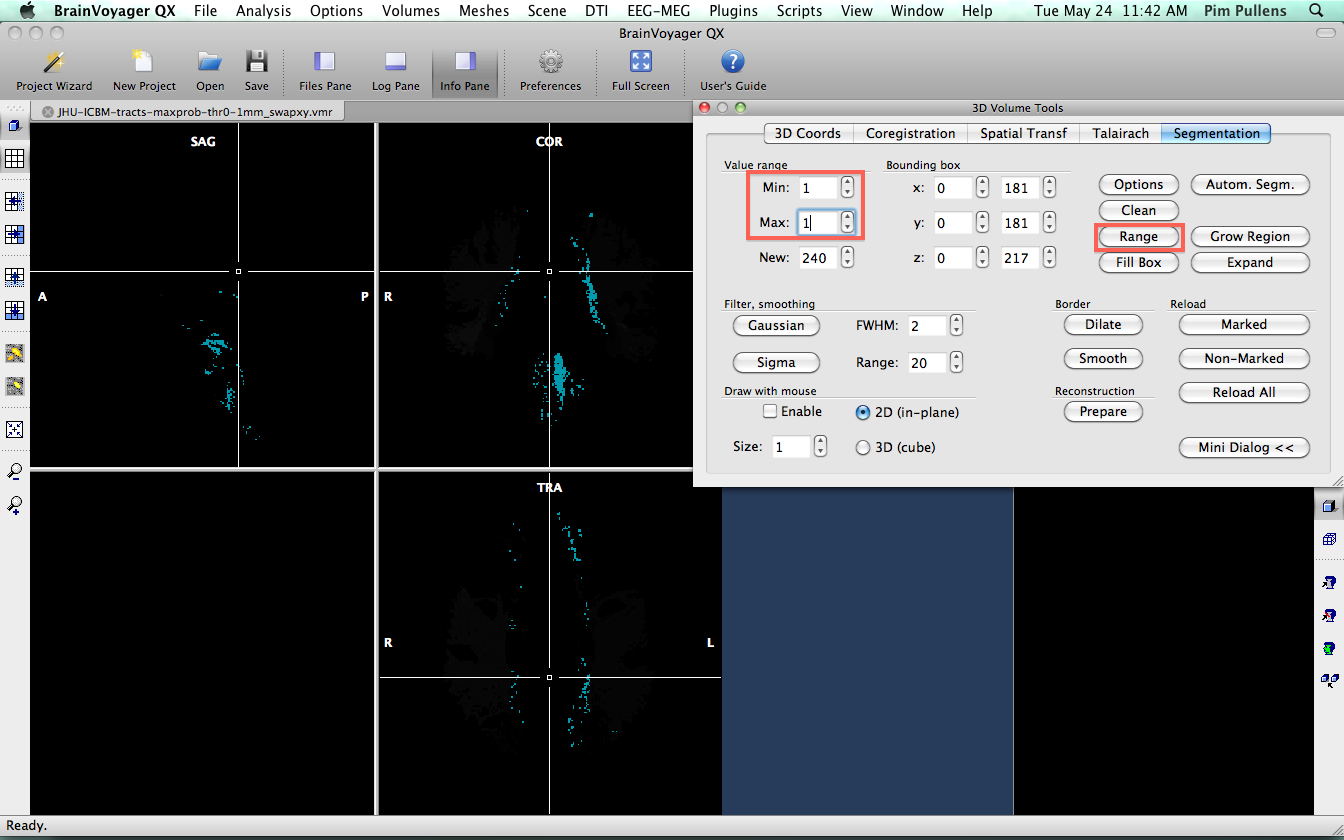
We know from the xml file that came with the atlas, that fibers labeled as "1" are part of the Anterior thalamic radiation L. So, we need to segment all voxels with value "1" and convert them into a VOI. Go to 3-D volume tools > segmentation. We want the voxels with value "1", so set lower and upper limit to "1" and click "Range" to get the voxels.
Next, click "Options" and "Define VOI". Name the voi "Anterior Thalamic Radiation L" and save. Go back to segmentation and click "Reload All" to refresh the VMR data and remove segmented voxels.
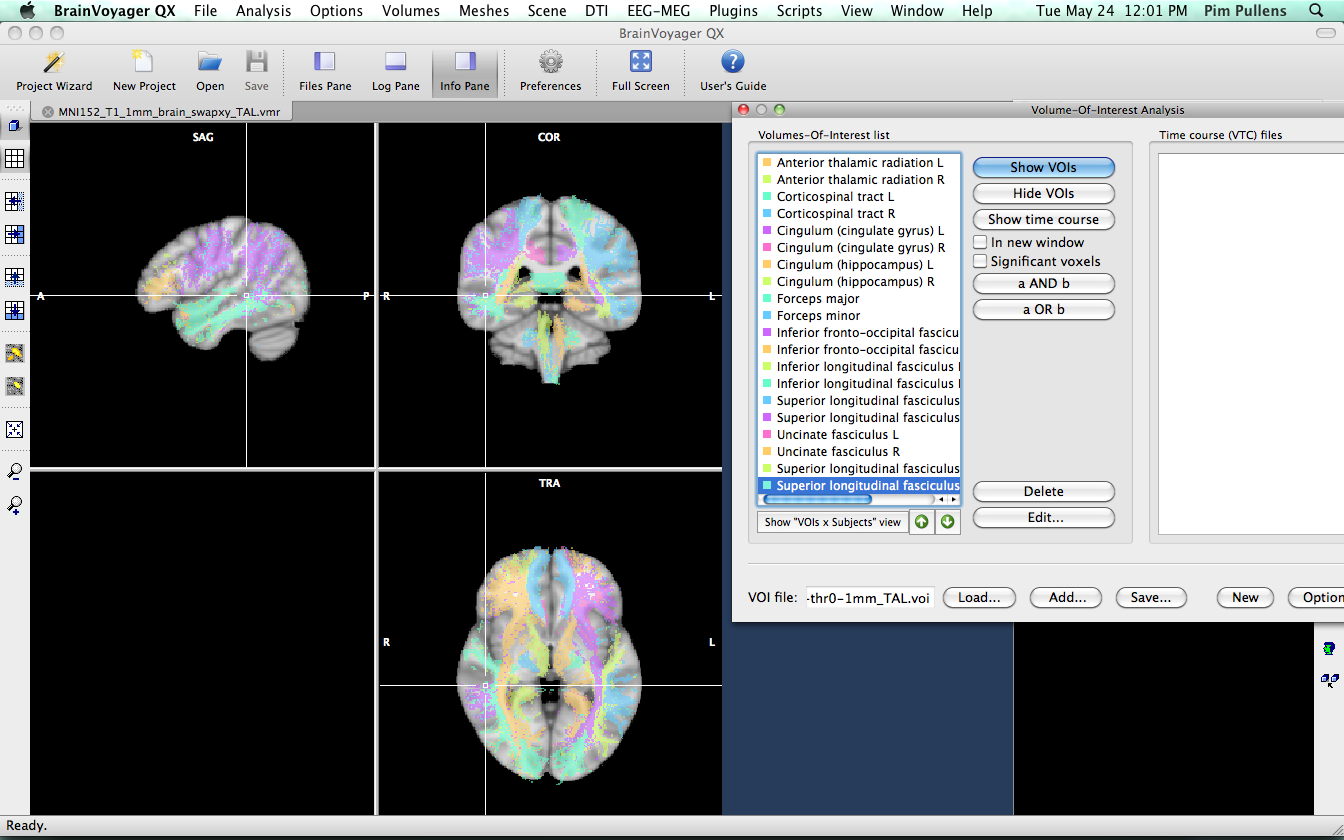
Repeat this process for all intensities 2...20 and name the VOIs according to the values found in the xml file. Below the result.