Creation of Physiological Noise Regressors Using External Recordings (Python Script)
- Details
- Category: Python Tools
- Last Updated: 19 July 2022
- Published: 23 May 2022
- Hits: 1402
Physiological fluctuations from heartbeat and respiration are a common source of noise in fMRI data.
Here we provide a Python script (CreatePhysioPredictors_BIDS.py) that will use pulse and/or respiratory recordings, saved in BIDS compatible format (*_physio.tsv.gz and _physio.json) (see https://bids.neuroimaging.io/ for more information on BIDS) as well as a corresponding BrainVoyager *.fmr file (created with BrainVoyager 21.4 and newer) to generate a set of volume-based physiological noise regressors.
Dependencies:
This script was developed with Python 3.8 and the following Python packages installed:
- NumPy (1.21.5)
- Scipy (1.4.1)
- Pandas (1.4.1)
- Matplotlib (3.5.1)
- Seaborn (0.11.2)
- PyQt (5.9.2)
- Json (2.0.9)
- Brainvoyagertools (0.2.0)
Download:
CreatePhysioPredictors_BIDS.py (version 0.1.0)
BatchProcessingPhysioFiles.py (version 0.1.0)
ExampleDataSet
Please note:
The BatchProcessingPhysioFiles.py can be used when multiple functional runs and their corresponding physiological datasets should be processed at the same time. It will ask for the script CreatePhysioPredictors_BIDS.py and for a TSV file containing the path information of the input files as specified below. For more details about the TSV file, please refer to the short description within the script BatchProcessingPhysioFiles.py.
However, please note that we strongly advise to use the batch processing only after you have familiarized yourself with the CreatePhysioPredictors_BIDS.py script. Furthermore, and even more important, please always check the input data quality for every dataset, since the generated noise predictors are only useful when the physiological raw data is of sufficient quality!
To get a first impression feel free to download and work with our example data set.
This is only a first version of the script; any suggestions, improvements and/or bug reports are highly appreciated and can be sent to This email address is being protected from spambots. You need JavaScript enabled to view it.
Documentation CreatePhysioPredictors_BIDS.py
A short description of the script
The script imports physiological recordings that follow the BIDS standard together with a corresponding FMR file. The physiological data, in form of the cardiac signal derived from a PPU (peripheral pulse unit) and the respiratory signal derived from a breathing belt, will be preprocessed, plotted and used for peak detection and the extraction of different physiological variables (e.g. heart rate, breathing rate, respiratory volume per time, etc.). Different study design matrices (SDM files) will be saved to disk, containing: (1) the filtered volume-based physiological signals (*_resp_filtz.sdm, *_cardiac_BP0.5-8Hz_z.sdm), (2) RETROICOR regressors, created based on the method proposed by Glover et al., 2000 (*_resp_RETROICOR.sdm, *_cardiac_RETROICOR.sdm, *_cardiacresp_RETROICOR.sdm), (3) and a set of physiological noise regressors, including the (shifted) heart rate (*_HR.sdm), the HR convolved with the cardiac response function (_*_HRCRF.sdm), the breathing rate (*_BR.sdm), the respiratory flow (*_RF.sdm), the (shifted) envelope of the respiratory signal (*_ENV.sdm), the (shifted) respiration variation (*_RV.sdm), the (shifted) respiration volume per time (*_RVT.sdm), RV convolved with the respiratory response function (*_RVRRF.sdm), and RVT convolved with the RRF (*_RVTRRF.sdm). If a (task) design matrix has been provided by the user, Pearson correlations of the task design with the derived physiological regressors will be computed, plotted and saved to disk. Additionally, if a stimulation protocol, specifying the experimental timing of the task, has been specified by the user, the stimulation protocol (PRT) together with the computed heart rate and/or breathing rate will be plotted and the mean HR and BR per event will be saved to disk.
Input Files
- Physiological Recordings:
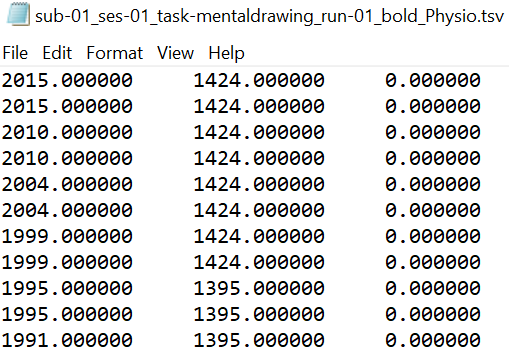
One pair of *_physio.tsv.gz and _physio.json files for one functional scan.
Both files should use the suffix "_physio". The compressed tsv file contains the continuous physiological recordings (cardiac and/or respiratory data) without header lines and with the last column representing the scanner trigger signal (coded with 0 and 1). The json file contains the following meta data: SamplingFrequency in Hz of all the data in the recording, StartTime in seconds in relation to the acquisition start of the first functional volume, name of Columns in the compressed tsv file.
Importantly, as specified by the BIDS standard: "Recordings with different sampling frequencies and/or starting times should be stored in separate files." In case of separate respiratory and cardiac files, both sets of files should be selected at the same time. The following naming scheme would be beneficial: sub-id_ses-id-task-name_run-id_recording-respiratory(or cardiac)_physio.tsv.gz.
If you have physiological signals from SIEMENS advanced physiological log or DICOM files, you can use the script SavePhysiologInBIDSfrom_readCMRRPhysio.py together with the script https://github.com/CMRR-C2P/MB/blob/master/readCMRRPhysio.py (from Marcel Zwiers) to convert these data traces to the BIDS standard.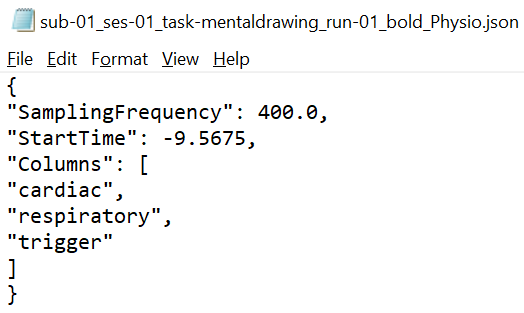
- fMRI Data:
The corresponding functional document of BrainVoyager (*.fmr). This file must have been created with BrainVoyager 21.4 or newer and will be used to extract and compare important scanning parameters. - Timing Information of the Experimental Task (Optional):
A corresponding stimulation protocol file (*.prt) can be loaded. This file will be used to plot and save the mean heart- and/or respiratory rate per stimulus condition and per event to disk. - A Single-Study Design Matrix (Optional):
A corresponding single-study design martix (*.sdm), containing the regressors of the task design or any confound regressors that the user would like to correlate with the derived physiological noise regressors. These correlations will be plotted in a correation matrix and saved to disk.
Output Files
All resulting files will be saved in a directory called 'PhysioOut', which is created in the same folder as the specified FMR file. All created files will have the same base filename as the FMR file followed by _*.
- Images of the Physiological Data Traces and Some Derived Noise Regressors:
Two PNG files showing the cardiac and respiratory signal, the identified peaks, the functional scan time and some of the derived physiological noise regressors.
*__cardiac.png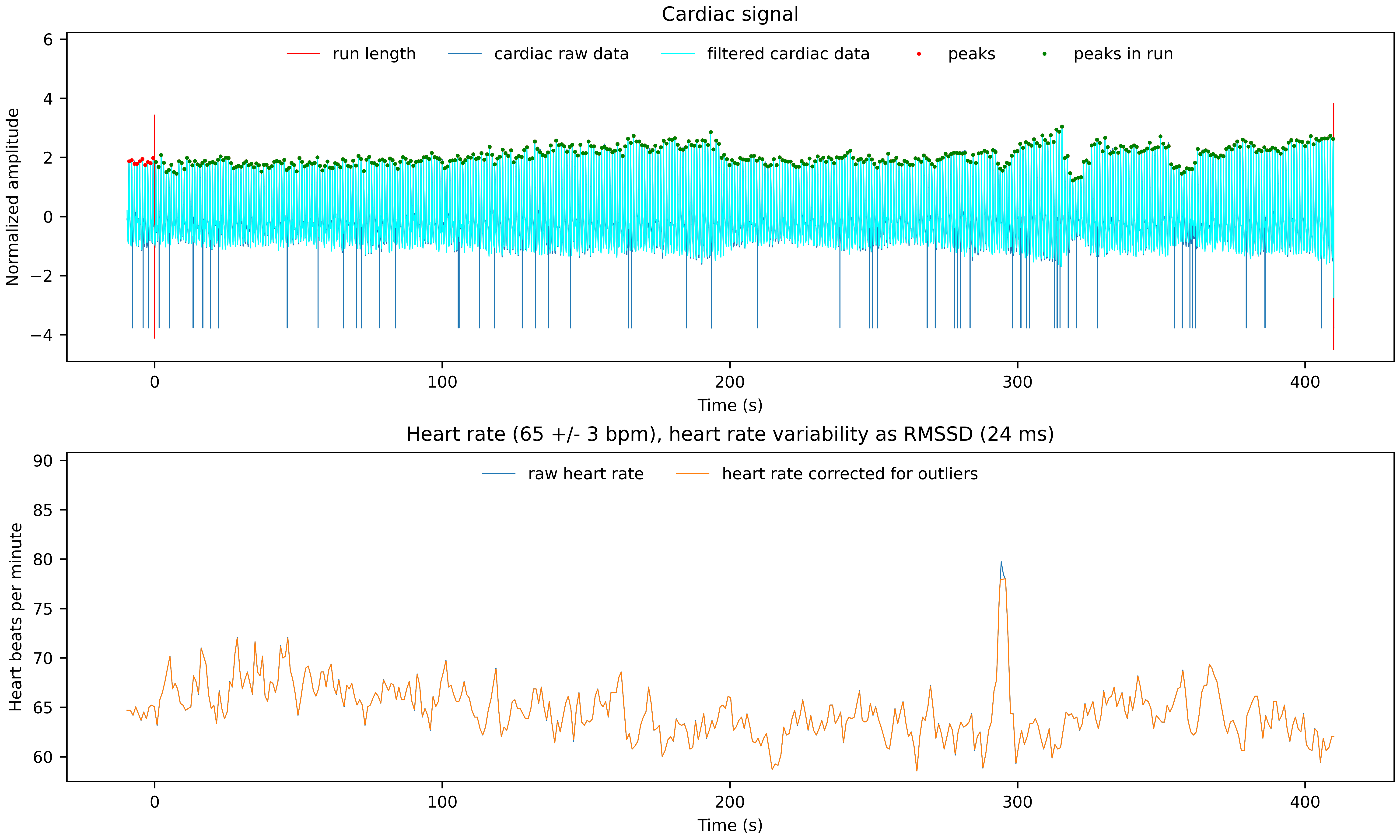
*_resp.png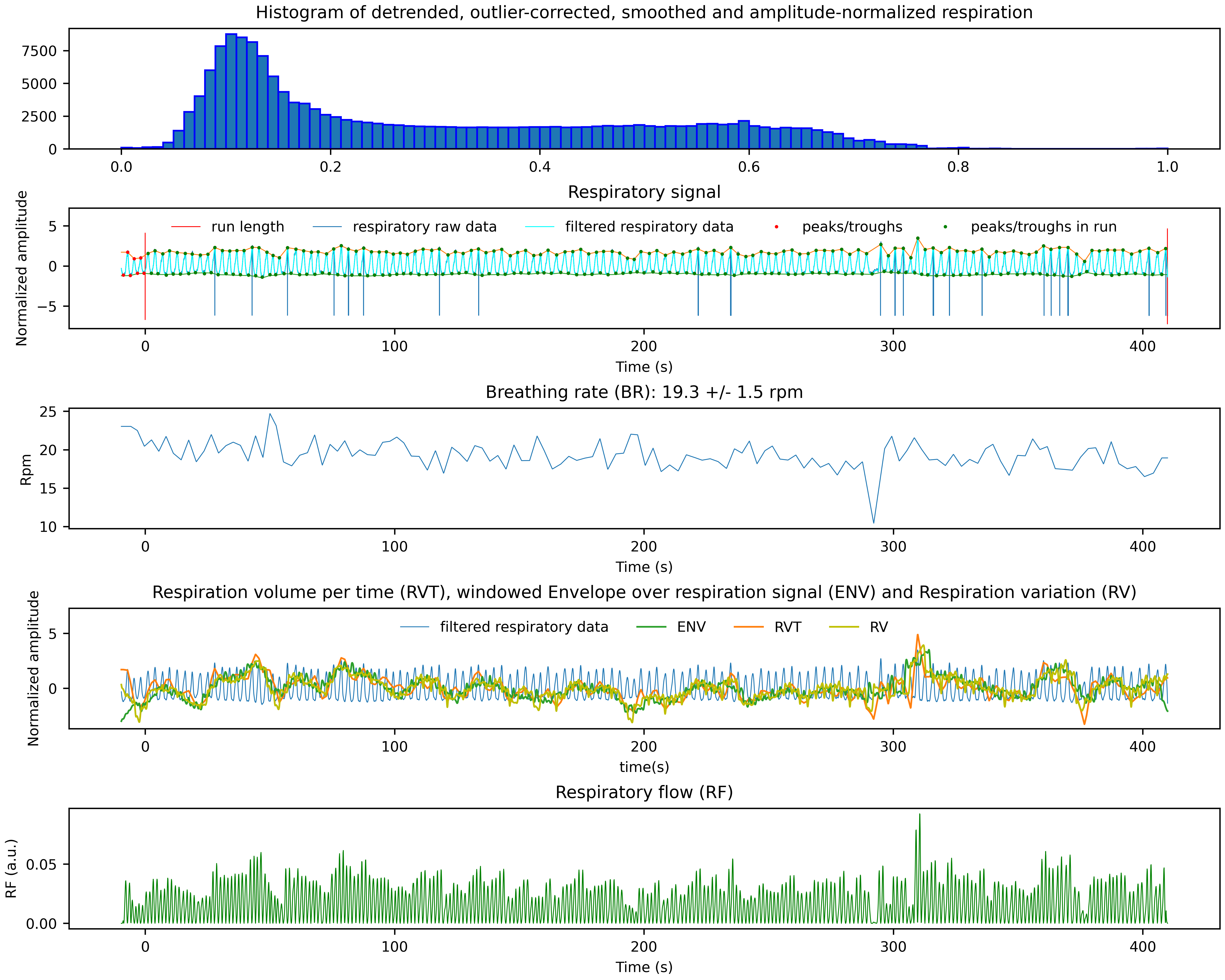
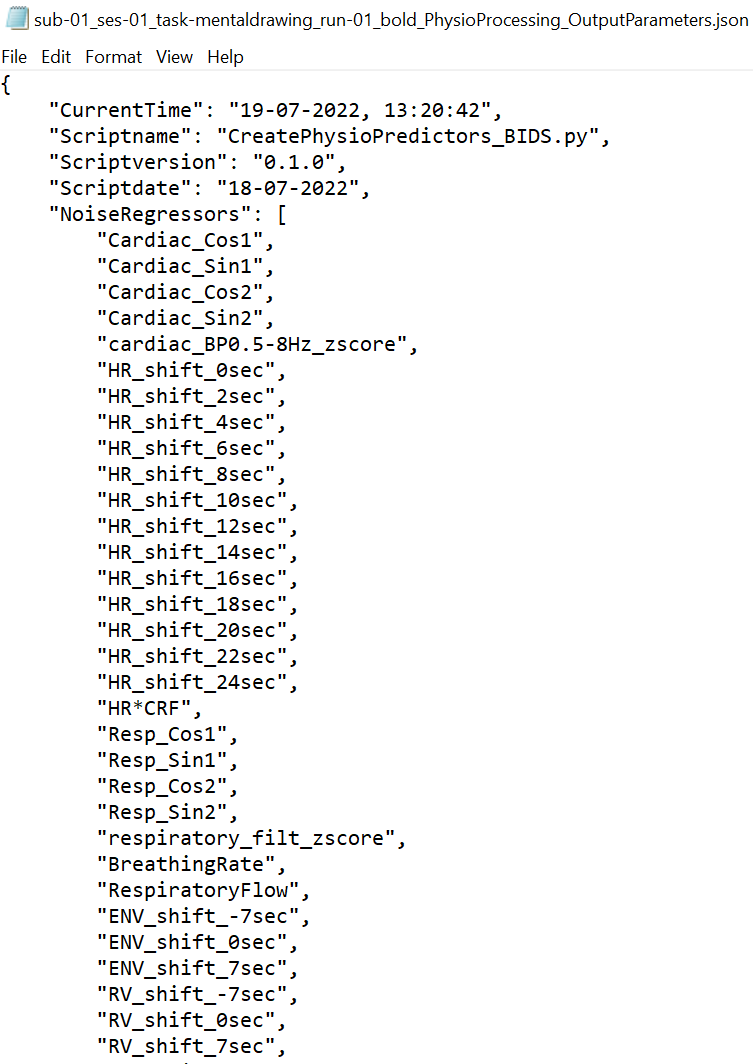
- Input and Output Parameters:
The Input, output and processing parameters will be saved in JSON files (*_InputParameters.json, *_OutputParameters.json)
-
Resulting Physiological Noise Regressors :
Separate design matrix files that can be used for physiological noise correction of the fMRI data will be saved to disk, including *_cardiac_BP0.5-8Hz_z.sdm, *_resp_filtz.sdm, *_cardiacresp_RETROICOR.sdm, *_HR.sdm, *_HRCRF.sdm, *_RV.sdm, *_ENV.sdm, *_RVT.sdm, *_RVTRRF.sdm, *_RVRRF.sdm). One TSV file with all physiological regressors will also be saved *_PhysiologicalNoiseRegressors.tsv - Task x Physiological Noise Correlation:
Provided that the user specified a task design matrix files as input, a task x noise correlation matrix with the corresponding p-values is saved as *_CorrelationMatrixNoiseTaskRegressors.tsv and as *_TaskNoiseCorr.png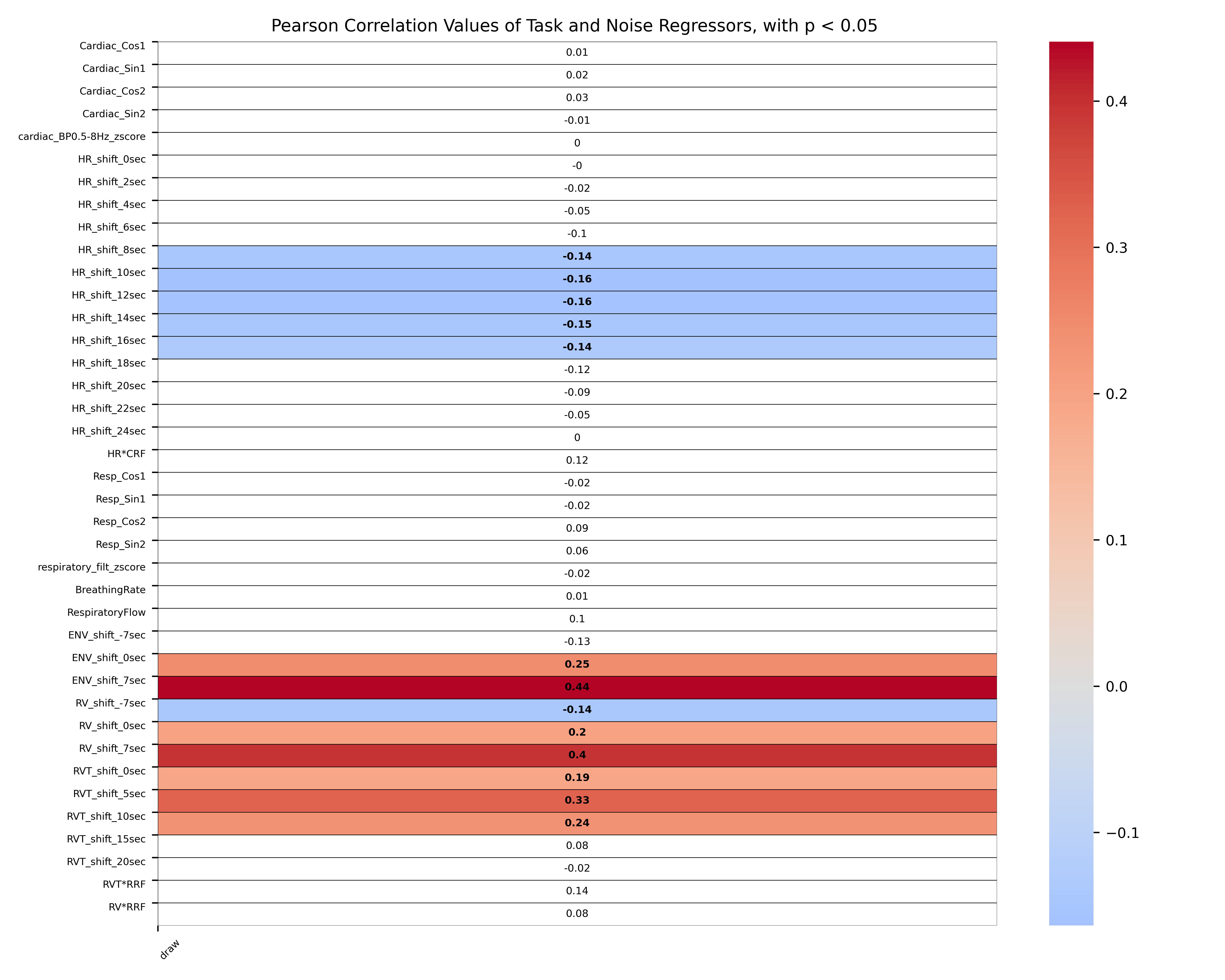
- Plotting the Experimental Design and the Heart- and/or Respiratory Rate of the Participant:
Provided the user specified a *.prt file with the experimental timings of the functional run, the stimulation protocol together with the heart- and/or respiratory rate will be plotted and saved as *_PRT_PhysioRate.png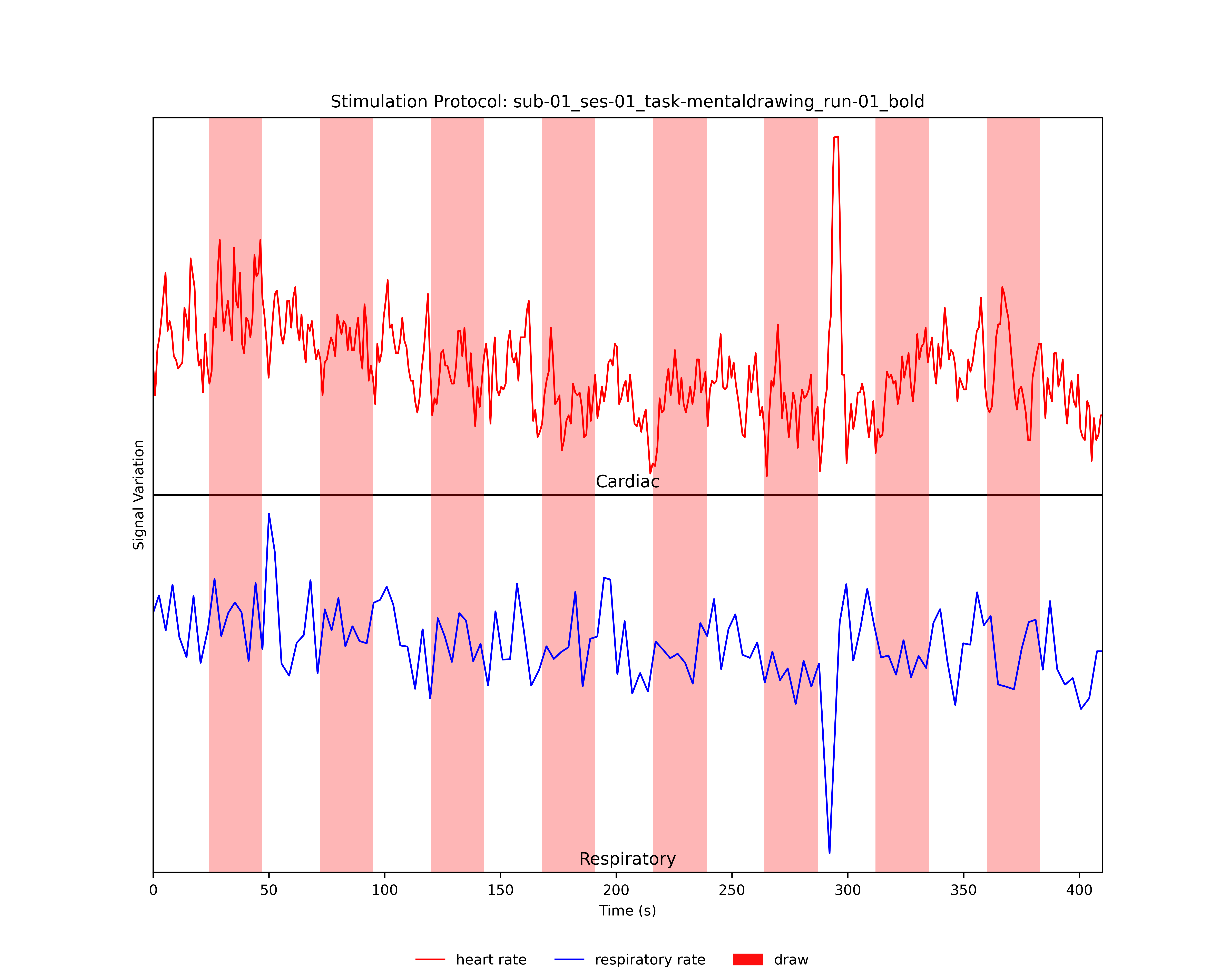
User-Specified Parameters in the Script
The section "user-specified parameter" in the script allows to adapt some input, processing and output parameters. Most values are set to reasonable defaults and don't need to be changed. References to relevant papers can be found in the comments of the script (next to the specific parameters).
physio_1file = True
- this parameter specifies if cardiac and respiratory raw data are saved together in a single JSON/TSV.GZ file. If only one of these physiological recordings exists or both recordings are saved together in a single file please set this parameter to True
cardiac_low = 0.5
cardiac_high = 8
- these two parameters specify the cutoff frequencies in Hz for the zero-phase second-order bandpass butterworth filter applied to the cardiac signal during the preprocessing
order_cardiac = 2
order_resp = 2
order_cardresp = 0
- These three parameters specify the number of harmonics for the definition of the RETROICOR model
- The effects of the cardiac and respiratory cycles are estimated as a linear combination of sinusoidal signals, the default order of correction terms for studies on brainstem imaging is c3r4i1 (e.g., 3,4,1, see Harvey et al., 2008) and can be reduced for other fMRI studies (e.g. to 2,2,0, see Power et al., 2017)
outlier_cardiac_threshold = 30
nIBIs = 5
- these two parameters are used to check the quality of the pulse peak detection of the cardiac signal
- inter-beat-intervals (IBIs) that differ by more than 30 percent (outlier_cardiac_threshold) from the mean of their 5 neighboring IBIs (nIBIs) are considered outliers
- if outliers are detected in the IBIs, the user can decide to test an alternative pulse peak detection for the cardiac signal
min_hbi = 0.60
- for the alternative pulse peak detection method, a minimum time interval between successive heartbeats (min_hbi in seconds) is assumed
- normal values vary from 0.40 to 0.90 seconds (values depend on age, gender, health, level of training, physical and emotional state)
hr_filloutliers_window = 25
hr_filloutliers_threshold = 10
- these two values are used for the outlier removal within the computed heartrate signal (based on Kassinopoulos, M., & Mitsis, G. D., 2019)
- the user needs to check whether these sudden changes (outliers) are indeed noise or true sudden changes in the cardiac signal
- the window size (hr_filloutliers_window) is defined in seconds
- the threshold (hr_filloutliers_threshold) defines the median absolute deviations (MAD) that are still within the normal range, all values above that threshold are considered outliers (normal values range from 3 - 20)
hr_shifts = np.arange(0, 25, 2)
- time shifts of the resulting heart-rate regressor that are saved within a design matrix (SDM)
- based on Shmueli et al., 2007
- in this example 13 versions of the heart-rate regressor are created, each shifted 2 seconds and ranging from no temporal shift (0 seconds) to shifting the predictor 24 seconds
resp_filloutliers_window = 0.25
resp_filloutliers_threshold = 3
- these two values are used for spike detection within the respiratory signal (based on Power et al., 2020)
- the window size (resp_filloutliers_window) is defined in seconds
- the threshold (resp_filloutliers_threshold) defines the median absolute deviations (MAD) that are still within the normal range, all values above that threshold are considered outliers (default is 3)
rvt_shifts = np.arange(0, 21, 5)
- time shifts of the respiratory volume per time regressor that are saved within a design matrix (SDM)
- based on Jo et al., 2010
- in this example 5 versions of the rvt regressor are created, each shifted 5 seconds and ranging from no temporal shift (0 seconds) to shifting the predictor 20 seconds
rv_shifts = np.arange(-7, 8, 7)
- time shifts of the respiration variation regressor that are saved within a design matrix (SDM)
- based on Power et al., 2017
- in this example 3 versions of the rv regressor are created, each shifted 7 seconds and ranging from shifting the predictor -7 seconds to +7 seconds
env_shifts = np.arange(-7, 8, 7)
- time shifts of the respiratory envelope regressor that are saved within a design matrix (SDM)
- in this example 3 versions of the env regressor are created, each shifted 7 seconds and ranging from shifting the predictor -7 seconds to +7 seconds
min_pvalue = 0.05
- this p-value for the heatmap of task-noise correlations is used to highlight all correlations between the physiological noise regressors and the task regressors that are statistically significant
