EPI distortion correction: echo spacing and bandwidth
- Details
- Category: Pre-Processing
- Last Updated: 09 April 2018
- Published: 09 April 2018
- Hits: 18259
EPI distortion correction - to correct for geometric distortions caused by the susceptibility artifact - in BrainVoyager can be performed as a preprocessing step with the anatabacus plugin. The anatabacus plugin uses the pixelshift method by Jezzard & Balaban (1995). The pixelshift for each voxel is calculated using a B0 fieldmap and some acquisition parameters: phase encoding bandwidth and phase encoding direction of the EPI data, and echo time difference of the "fieldmap" [1].
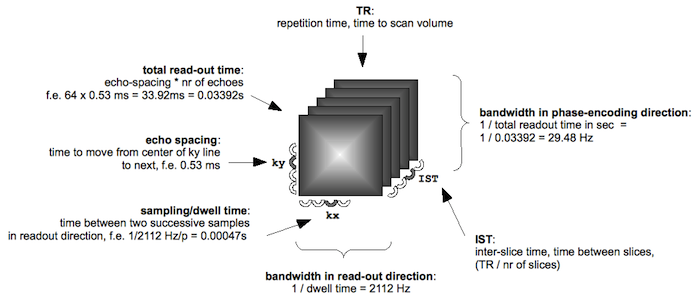
Figure: acquisition parameters; please note that FSL uses the word 'dwell time' in the sense of 'echo spacing'.
To calculate the bandwidth (BW) in phase encoding direction, we need to know the echo spacing (ES). Echo spacing is related to the receiver bandwidth BW (the range of frequencies that can be captured with a certain sampling speed) as follows: ES = 1/BW.
Echo spacing for different scanners
Siemens: the echo spacing in milliseconds is mentioned on the scan protocol PDF. For details, please consult the anatabacus manual.
echo spacing in msec = 1000 * (water-fat shift (per pixel)/(water-fat shift (in Hz) * echo train length))
- echo train length (etl) = EPI factor + 1
- water-fat-shift (Hz) = fieldstrength (T) * water-fat difference (ppm) * resonance frequency (MHz/T)
- water-fat difference (ppm) = 3.35 [2]
- resonance frequency (MHz/T) = 42.576 (for 1H; see Bernstein pg. 960)
[1] For Siemens scanners, the fieldmap sequence is gre_field_mapping. This sequence acquires two similar T2 images with different echo times. Magnitude and phase images are reconstructed (usually one only receives the magnitude data). Since the two phase images are subtracted from each other, the result is two magnitude and one so-called 'differential phase map'. After unwrapping (removing jumps of 2 pi) and rescaling to Hertz, this differential phase map is used to calculate the shift for each pixel.
[2] Haacke et al: 3.35ppm. Bernstein et al (pg. 960): Chemical shifts (ppm, using protons in tetramethyl silane Si(CH3)4 as a reference). Protons in lipids ~1.3, protons in water 4.7, difference: 4.7 - 1.3 = 3.4.