This section is experimental: please check the fiber orientation carefully after the conversion!
In this page is described how to convert TrackVis (www.trackvis.org) trk files into BrainVoyager fiber files (*.fbr).
Prerequisites
- The m-files needed for the conversion were tested with Matlab R2008a.
- BVQXtools or NeuroElf, to download from www.neuroelf.net
- TrackVis read/write functions to download from https://github.com/johncolby/along-tract-stats
- Add BVQXtools and TrackVis read/write functions to your Matlab path
- Download the following file and put the unzipped m-file into your Matlab path:
trk_to_fbr.zip
Getting Started
- Create DMR and VMR in BrainVoyager
- Co-register DMR to VMR, create VDW and DDT and make sure the tensors display correctly
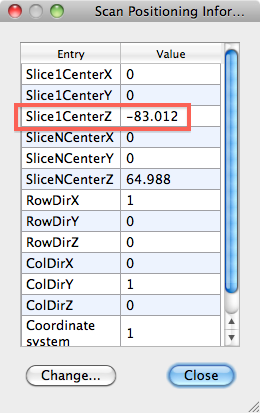
- From the DMR, get the POS info as displayed below (in DMR Properties > Pos info), in this example -83.012
- From the VMR get the POS info (in VMR Properties > Pos info), in this example -11.5663
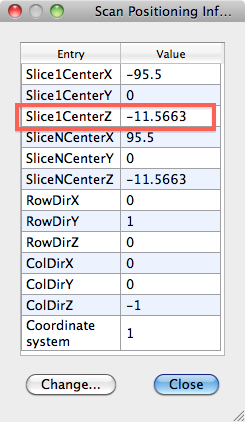
- Subtract the absolute DMR pos value from the absolute value of the VMR pos info, i.e. 83.012-11.5663 = 71.4457. Write this value down.
- For the conversion, we need the offset from the TrackVis origin to the BrainVoyager origin. For the x and y offset, this is usually 0.5*X/Y dimension of the VMR, so 256/2=128 in most cases. The z offset is the value we calculated earlier, i.e. 71.4457. These 3 values are inputs to the conversion function.
In Matlab, do
>> cd /path-to-TrackVis fibers/
>> fbr = trk_to_fbr(128,128,71.4457); % this is trk_to_fbr(XOffset,YOffset,ZOffset)
The program will first ask for a trk file, and then to save the fbr file.
Example
| TrackVis 0.5.1 | BrainVoyager 2.3.0 |
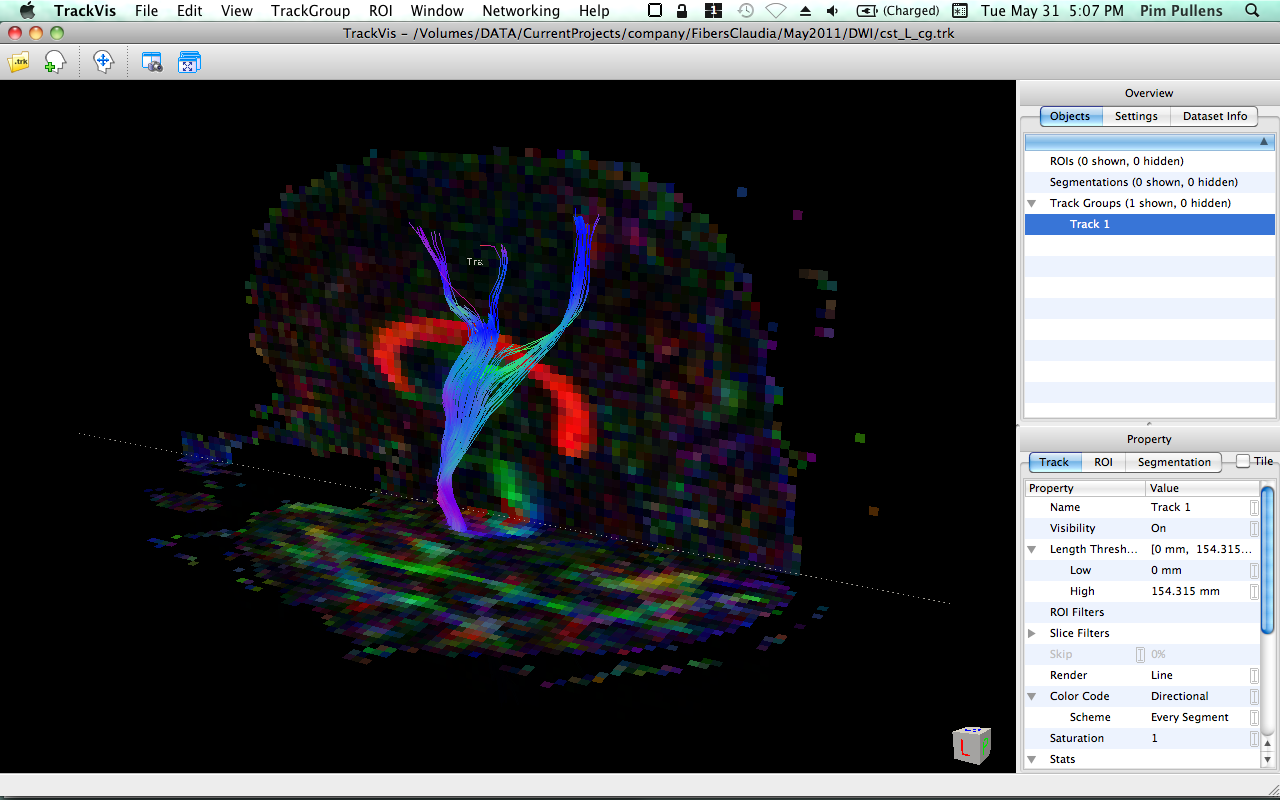 |
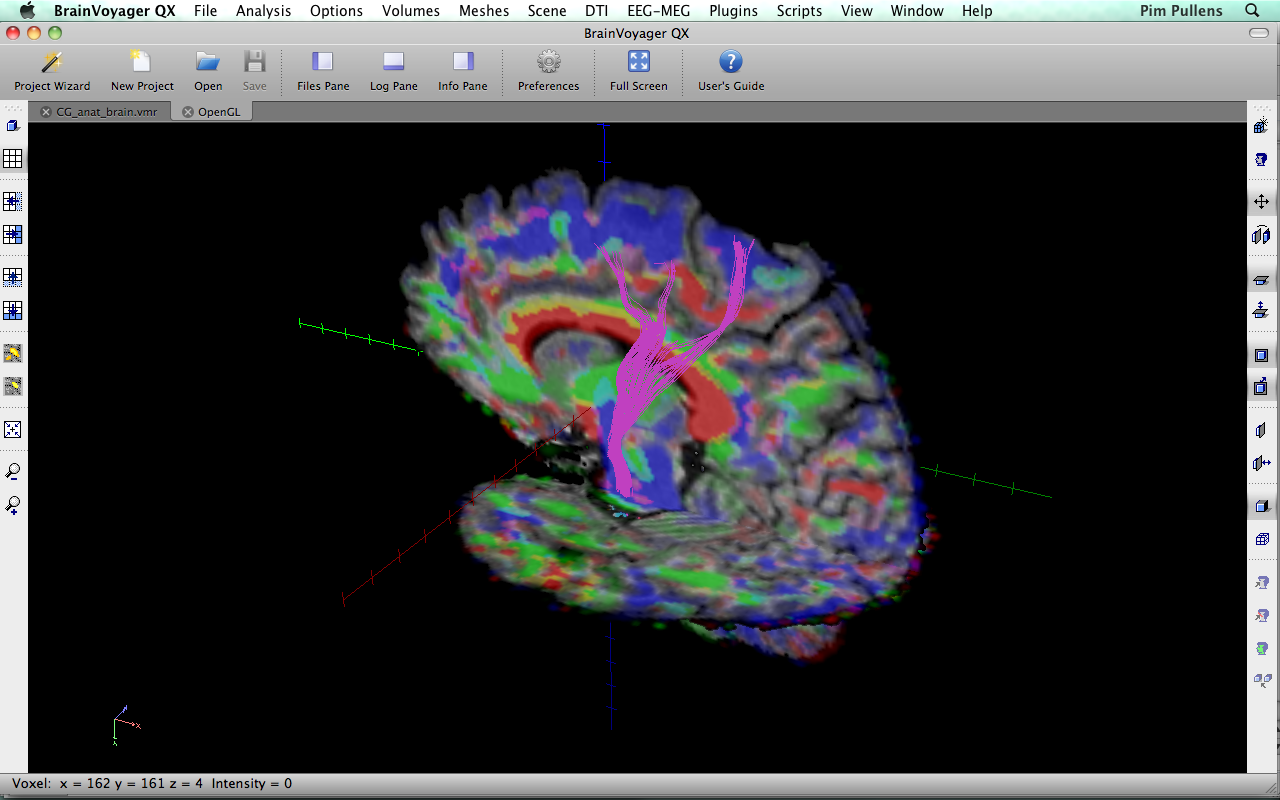 |