Head motion is probably the most severe but also an unavoidable problem in fMRI studies. Of course all subjects participating in an fMRI study are instructed not to move, however, it's impossibe to lie completely still during a time interval of 1 to 2 hours. Everybody who has been at least once in an MRI scanner is aware of that fact.
Nevertheless, this is a severe problem for statistical analysis of the data, since it is assumed that each voxel represents one unique location in the brain. If the subject however moved, the time course of one single voxel would represent a signal derived from different parts of the brain. In extreme cases, those effects might be visible in “ring activations” around the edges of the brain resulting from the fact that the intensity differences between adjacent voxels are particularly high at tissue boundaries.
To minimize those false positive activations while increasing sensitivity to true task-related activations, correction algorithms have to be employed to correct for motion artefacts.
In general motion correction incorporates the estimation of the movement based on one reference volume and the subsequent application of the estimated movement parameters to the data to realign the time series of brain images to the reference. The realignment is applied by rigid body transformations, which assume that the shape and size of the to be coregistered volumes are the same and that one image can be spatially matched to another one by the combination of three translation (in mm) and three rotation parameters (pitch, roll, yaw; in degrees). In the movies below, you can see for each parameter the respective movement direction and the plotting of the motion estimate.
Translation X:
Translation Y:
Translation Z:
Rotation X (pitch):
Rotation Y (roll):
Rotation Z (yaw):
Those motion estimates are visualized in a similar plot in BrainVoyager while motion correction is performed (see figure below).
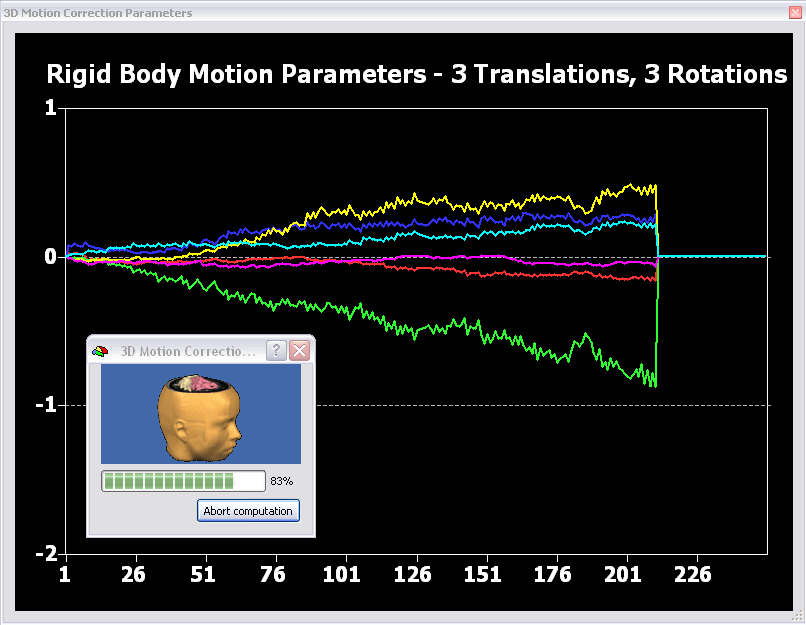
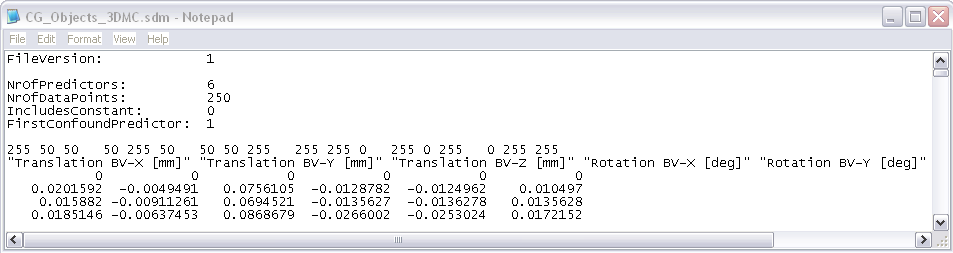
In addition to the plot and a motion-corrected data set (“_3DMC.fmr”), there are also two files saved to disk containing the motion estimates: “_3DMC.log” and “_3DMC.sdm”. While the log-file can be used to inspect the values of the parameters, the sdm-file can be used to add the motion parameters as confounds to the general linear model since they are saved in the format of a design matrix.
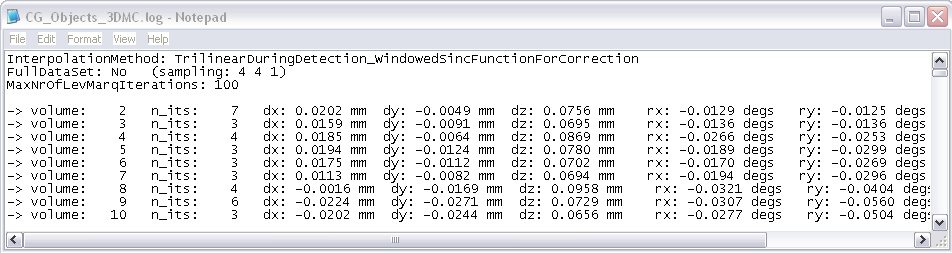
To save more detailed information about the motion estimates for each iteration step, check the “Create extended log file” option which results in an additional log file ending with “_3DMC_verbose.log”.
There are different options available in BrainVoyager QX allowing to adapt the method to your requirements. First of all you can choose between three different interpolation methods: trilinear, sinc, or a combination of trilinear for detection of motion and sinc for correction. The method used is also indicated in the resulting filename following the _3DMC with T for trilinear, S for sinc and TS for a combination of both. While the computation cost of sinc interpolation is very high, trilinear interpolation slightly smoothes the data spatially. Therefore, it is recommended to use the “Trilinear/sinc interpolation” option (default) to avoid the problem of inducing unwanted blurring effects in the data while preserving a reasonable computation time. Furthermore, it has been shown that it is not necessary to use all voxel values for estimation of the movement parameters. Hence, BVQX uses by default a reduced number of voxels which can be changed by deselecting the “Reduced data” option.
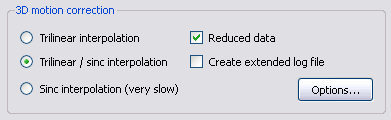
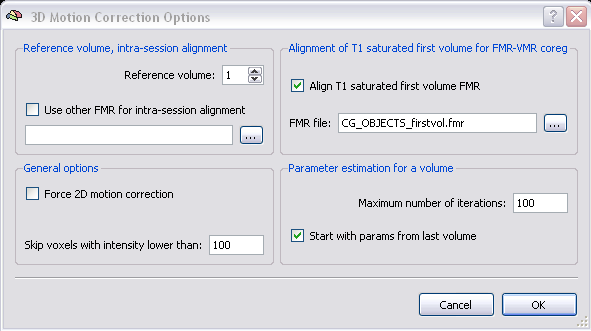
Even more parameters can be adapted in the “3D Motion Correction Options” dialog which can be evoked by clicking on the button “Options” in the “3D motion correction” field.
You can choose the reference volume for the dataset (default is the first volume) or even another FMR for alignment (of course acquired in the same session!). The last mentioned option would make sense, for example if another functional data set was acquired closer to the anatomical data (to improve the coregistration of both data sets later on). However, it is important to stress the point that this option is not meant for aligning different sessions, since it cannot easily correct big misalignments.
By default the T1 saturated first volume fmr is also aligned to the reference volume. The reason for that is that it is often recommended to use the first volume fmr for the coregistration of the functional and anatomical data set, since the first volume is characterized by a more pronounced contrast which improves the alignment of the data.
Note: Although the “_firstvol.stc” is changed by the motion correction procedure (when the default values are kept) it is not apparent in the file name!
There is also the possibility to correct for motion only in the x-y plane and to ignore the motion between slices by checking “Force 2D motion correction”. This is however only recommended, when the data set contains only a few slices (less than 5).
BrainVoyager masks out noise voxels by default using only voxels with an intensity above 100, when this value is set to 0 also voxels of the background are used for the motion correction process.
In the field “Parameter estimation for a volume” you can specify the maximum number of iterations used for each volume. This value is set to 100 which is very conservative and usually not necessary to estimate the motion parameters. Since head motion is usually incremental, meaning that the motion increases from volume to volume over time, the parameter estimates of the last volume are used by default to start the estimation process of the motion parameters for the current volume, you can uncheck this option when you expect rather sudden movements to dominate the functional data set.
There are different motion patterns which have to be taken into account: the first one is random motion, meaning that the movement of the subject is not correlated with the experimental paradigm; the second one on the other hand is task-correlated motion, which predominantely occurs in motor mapping experiments in which subjects are forced to move in response to certain stimuli (e.g., button presses or overt speech).
In BrainVoyager QX there are two different options to correct motion-related artefacts: motion correction as a preprocessing step alone or motion correction in preprocessing combined with inclusion of the realignment parameters as confounds of no interest in the general linear model.
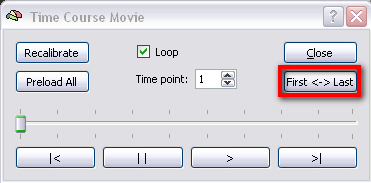
Of course not all kind of motion can be corrected via the rigid body alignment (especially sudden spikes), therefore it is important to ckeck the improvement of the data after motion correction by running the “Time Course Movie” tool in the “Options” menu, which displays the functional data across time. To easily detect motion, click the "First Last " button, which alternately shows the first and the last volume.
Note: This tool is also very useful to screen the data for other artefacts.
In the movies below you see on in the upper one the original data set with motion visible in the "Time Course Movie" and in the lower one the same data set after motion correction with reduced motion.
As a rule of thumb, functional data sets with motion greather than the voxel size should be discarded from analysis, since this amount of motion cannot be corrected properly by applying motion correction procedures (Formisano et al., 2005).
Plotting Motion using the *3DMC.sdm file
You can use the following maltlab script to create motion plots similar to the ones created by BrainVoyager: MotionEstimatesPlots.m
The advantage is that you can load multiple *3DMC.sdm files at once. The output will be a *3DMC_MotionPlot.png file saved in the same folder as the *3DMC.sdm file as well as a Motion.mat file including for each run the maximum motion with respect to a reference run, the maximum motion within the same run, as well as the maximum range of motion within the same run. In case the reference run is the same as the current run, the maximum motion with respect to a reference run should be the same as the maximum motion within the same run.
References:
Formisano E, Di Salle F, Goebel R. Fundamentals of data analysis methods in functional MRI. In: Landini L, Positano V, Santarelli MF, editors. Advanced imaging processing in magnetic resonance imaging. CRC Taylor & Francis, 2005. pp. 481-503.
Morgan VL, Dawant BM, Li Y, Pickens DR. Comparison of fMRI statistical software packages and strategies for analysis of images containing random and stimulus-correlated motion. Comput Med Imaging Graph 2007; 31: 436 - 446.