How to use the copy labels approach
- Details
- Category: Cortex Based Alignment
- Last Updated: 13 April 2018
- Published: 13 April 2018
- Hits: 4300
BV QX version used: 1.10.4 (1252)
Dataset used: CBA example dataset
It is very helpful to obtain automatic labelling of cortical structures in a specific surface reconstruction. BrainVoyager offers and option to use predefined patches of interest (POIs) in combination with cortex-based alignment to make optimal use of a couple of standard POIs coming with the standard installation.
When not including the cortex based alignment information into the POI labelling,
any predefined area – which is based on a specific anatomy - will not fit too well on another subject-specific cortex.
I.The problem:
1. To demonstrate the mismatch, we overlay to predefined POIs on a surface reconstruction:
In BVs standard installation, all the necessary information can be found in the so called “Atlas Brains” folder.
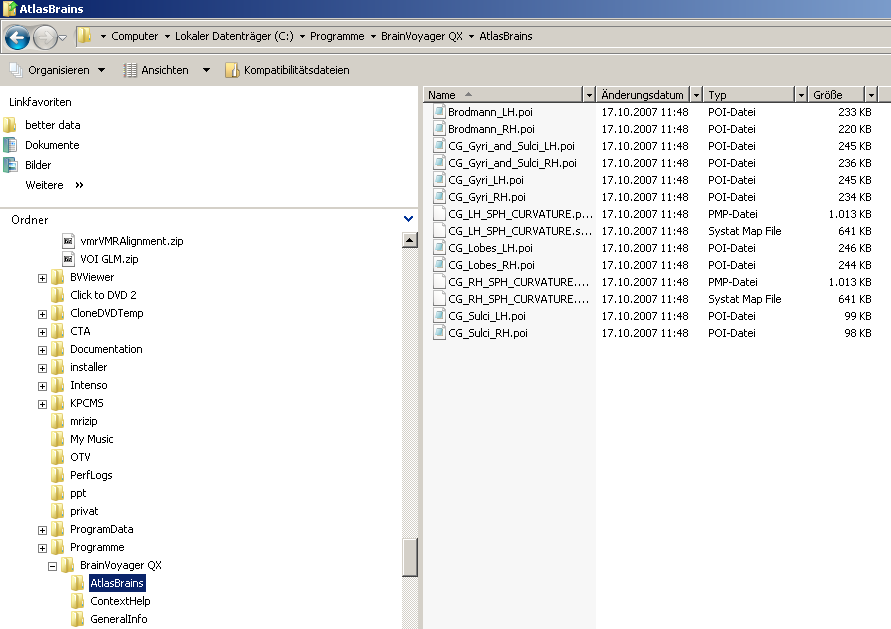
2. The folder contains several POI files as well as curvature maps necessary for the cortex based alignment.
To check the current status of overlap between the predefined POIs and a specific anatomy, we load an SPH mesh (it has to be a standard mesh, because the POIs have been defined on the basis of a standard mesh) and load one of the Atlas Brains folders POI files. In this case, we use subject “SG” of the cortex based alignment example dataset.
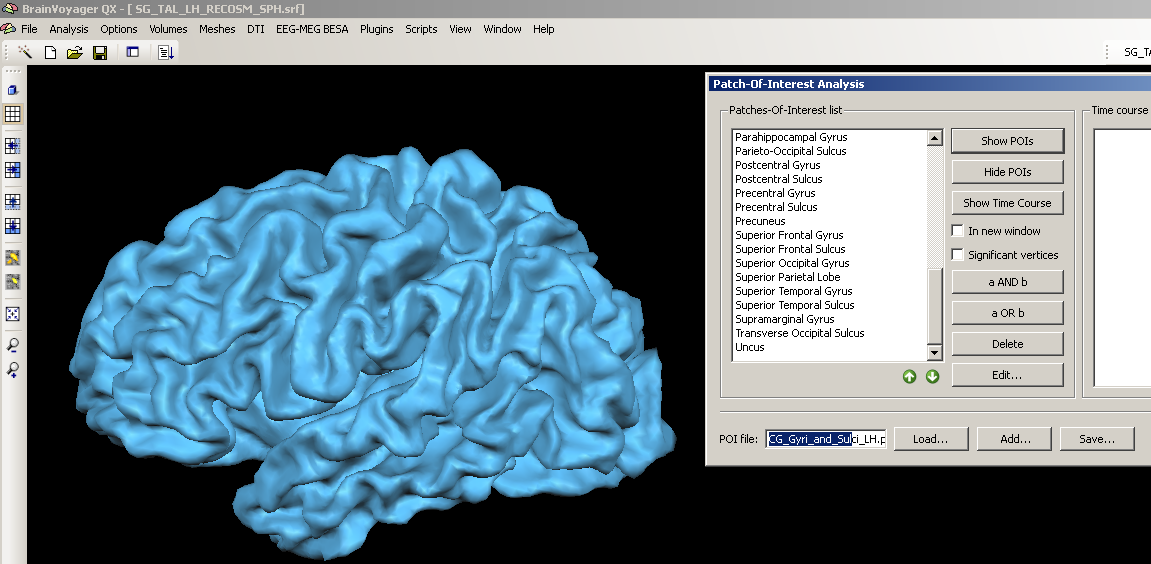
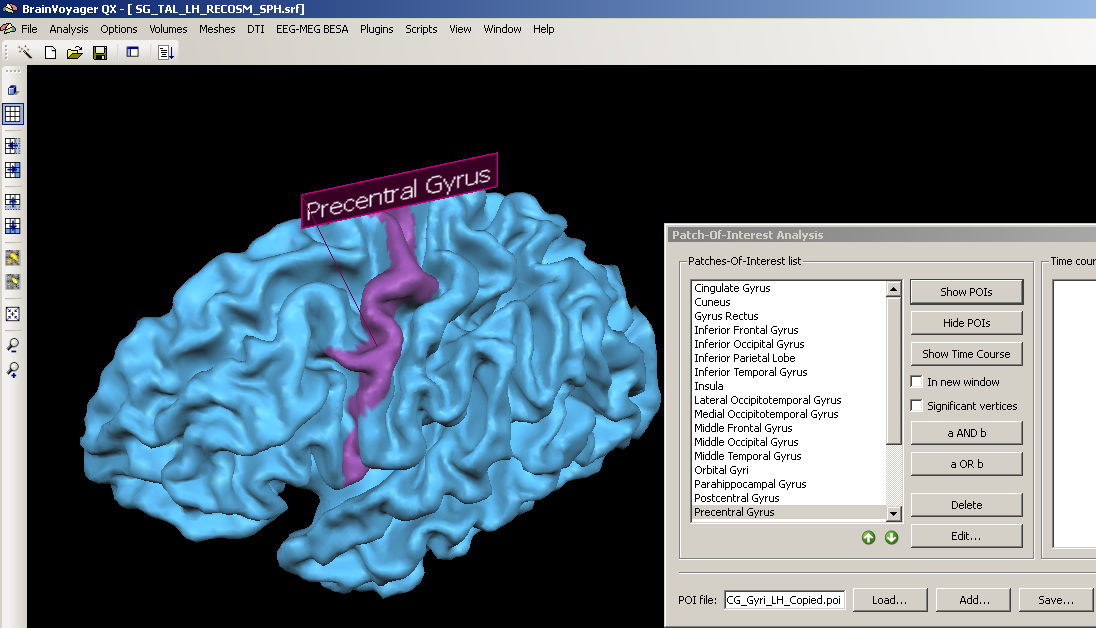
3. We choose a structure within the POI file, e.g. the precentral gyrus, clicking the “Show POIs” button, we display it on the currently created mesh.
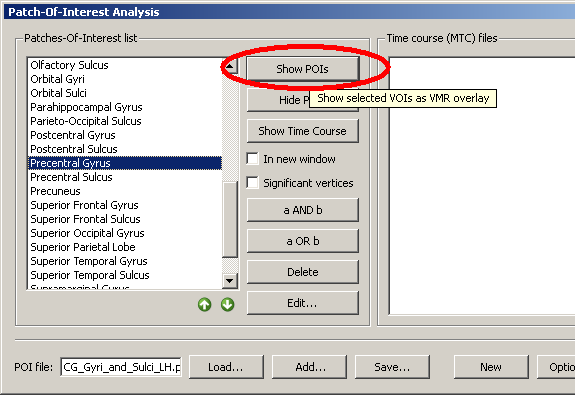
We can see clearly that the structure is not properly matched but that the demarcated area is too far anterior.
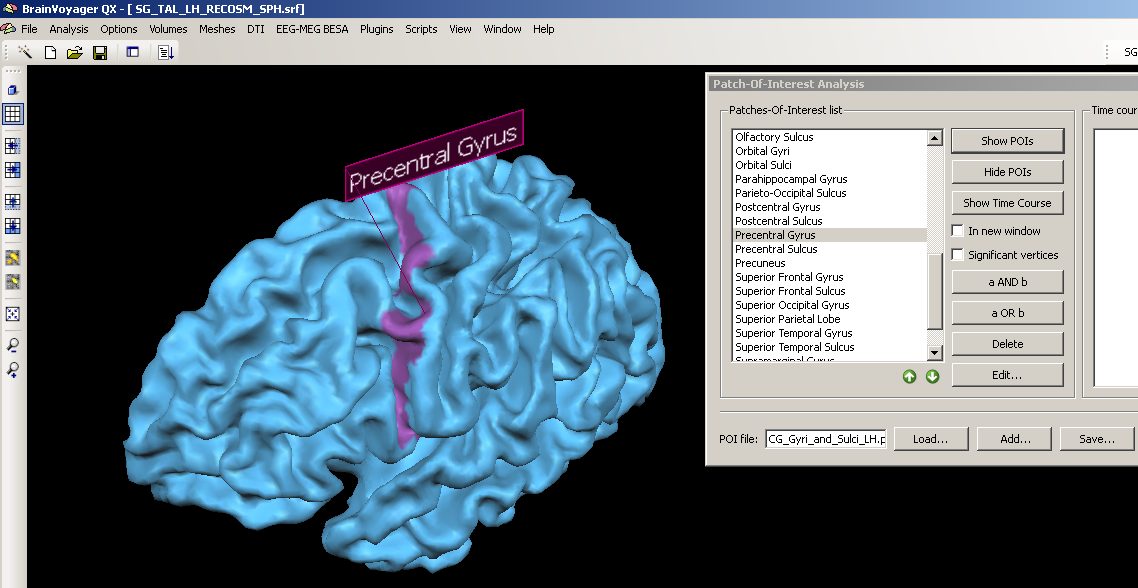
To optimally use the POI labels in the Atlas brains folder, we have to use the cortex- based alignment technique.
II. Using the cortex based alignment:
1. We load a hemisphere we would like to align to the atlas brain.
We open the left hemisphere representation of subject “SG” of the CBA example dataset.
2. We open the cortex based alignment dialog.
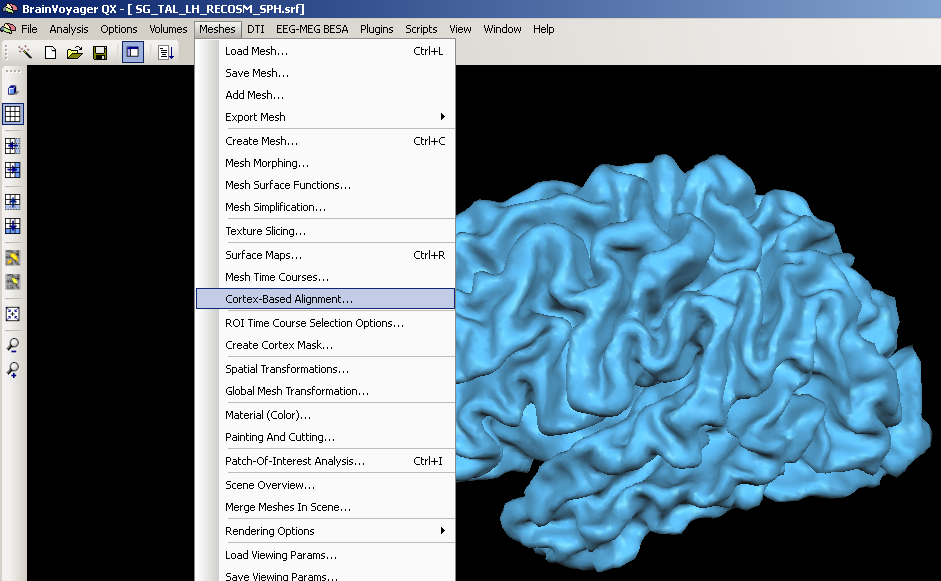
3. From here on, the further steps of processing depend on the current status of data processing. If the data has not yet been used in cortex-based alignment, one has to create and process the corresponding files first (Sphere, SPH file).
In this case, we have already prepared the data and may start directly with the alignment between source(s) and target.
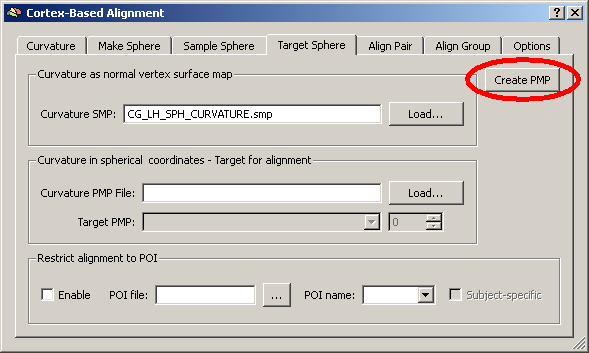
4. We open the “Target Sphere” tab. We load the prepared SMP file of the left hemisphere of subject “CG” (from the “Atlas brains” folder) and click the “Create PMP” button.
We open the “Align group” tab an used the “Add” button to Add the curvature SMP of subject “SG”. Below the list of files, we check the radio button “Align each entry to target sphere”.
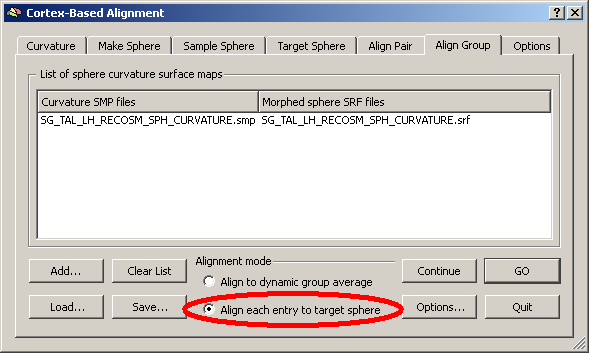
In the list of files to align, we can of course enter multiple curvature maps (left hemisphere curvature representations of different brains) that we can all align together.
After the alignment is performed, we open the Target Sphere tab and click the “Copy Labels…” button.
A new dialog opens up where we have to load:
a) the POI file we would like to display (using one of the Atlas brains folders POIs)
b) the source sphere mesh – this is SPH mesh of the dataset which is finally used to display the areas of interest.
c) the sphere-to-sphere mapping file, which has just been created during the aligment. Because we have used the CG curvature as the target of the CBA, we have to use the “INV.ssm” file.
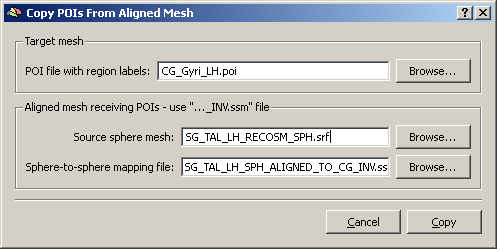
By clicking the “Copy” button, we create a new POI file on the basis of the old POI file and the cortex-based alignment result (SSM file). This is automatically opened up and called “CG_Gyri_LH_Copied.poi”.
We check again the position of the precentral gyrus by using the Show POIs button: